Biology:Phosphoribosylanthranilate isomerase
| phosphoribosylanthranilate isomerase | |||||||||
|---|---|---|---|---|---|---|---|---|---|
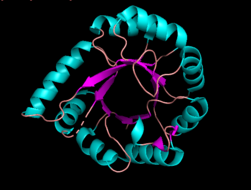
 3D rendering of Phosophoribosylanthranilate Isomerase | |||||||||
| Identifiers | |||||||||
| EC number | 5.3.1.24 | ||||||||
| CAS number | 37259-82-8 | ||||||||
| Databases | |||||||||
| IntEnz | IntEnz view | ||||||||
| BRENDA | BRENDA entry | ||||||||
| ExPASy | NiceZyme view | ||||||||
| KEGG | KEGG entry | ||||||||
| MetaCyc | metabolic pathway | ||||||||
| PRIAM | profile | ||||||||
| PDB structures | RCSB PDB PDBe PDBsum | ||||||||
| Gene Ontology | AmiGO / QuickGO | ||||||||
| |||||||||
In enzymology, a phosphoribosylanthranilate isomerase (PRAI) (EC 5.3.1.24) is an enzyme that catalyzes the third step of the synthesis of the amino acid tryptophan.[1]
This enzyme participates in the phenylalanine, tyrosine and tryptophan biosynthesis pathway, also known as the aromatic amino acid biosynthesis pathway
In yeast, it is encoded by the TRP1 gene.[2]
Nomenclature
This enzyme belongs to the family of isomerases, specifically those intramolecular oxidoreductases interconverting aldoses and ketoses. The systematic name of this enzyme class is N-(5-phospho-beta-D-ribosyl)anthranilate aldose-ketose-isomerase. Other names in common use include:
- PRA isomerase,
- PRAI,
- IGPS:PRAI (indole-3-glycerol-phosphate,
- synthetase/N-5'-phosphoribosylanthranilate isomerase complex), and
- N-(5-phospho-beta-D-ribosyl)anthranilate ketol-isomerase.
- xPRAI (monomeric variant in Saccharmyces cerevisiae)[3]
- PRAI[ML256-452] (engineered variant of 1-(2-carboxy-phenylamino)-1-deoxy-D-ribulose 5-phosphate carboxylase: PRAI)[3]
Reaction[4]
Phosphoribosylanthranilate isomerase is one of the many enzymes within the biosynthesis pathway of tryptophan (an essential amino acid). The upstream* pathway substrates and intermediates are shown below (Fig. 2).
As seen in Fig. 3, N-(5'-phosphoribosyl)-anthranilate via this enzyme is converted into 1-(o-carboxyphenylamino)-1-deoxribulose 5-phosphate. As the name phosphoribosylanthranilate isomerase suggests, it functions as an isomerase, rearranging the parts of the molecule without adding or removing molecules or atoms.
The reaction seen in Fig. 3, is an intramolecular redox (reduction-oxidation) reaction.[5] Its first step involves a proton transfer. This product intermediate, an enolamine, is fluorescent, which is useful for kinetic studies
within this pathway.[5] However, this product is unstable, and quickly isomerases into an α-amino keto.
- Note: Upstream/Downstream are relative to the compounds/molecules directly involved in phosphoribosylanthranilate isomerase reaction
Kinetics
Michaelis–Menten kinetics data, is given in the table below for PRAI and indole-glycerol-phosphate synthase (IGPS, EC 4.1.1.48).[6]
| Enzyme | Temperature (°C) | Km
(μM) |
kcat
(1/sec) |
|---|---|---|---|
| tPRAI | 25 | 0.280 | 3.7 |
| 45 | 0.390 | 13.5 | |
| 60 | 0.730 | 38.5 | |
| 80 | 1.030 | 116.8 | |
| tIGPS | 25 | 0.006 | 0.11 |
| 45 | 0.014 | 0.75 | |
| 60 | 0.053 | 3.24 | |
| 80 | 0.123 | 15.4 |
Structure
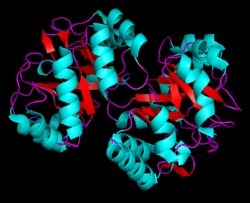
Depending on the microorganism PRAI's structure can vary between a mono-functional enzyme (monomeric and labile) or a stable bi-functional dimeric enzyme. Within Saccharomyces cerevisiae, Bacillus subtilis, Pseudomonas putida, and Acinetobacter calcoaceticus the enzyme is monmeric.[7] In contrast, in hyperthermophile Thermotoga maritima, Escherichia coli (Fig. 5), Salmonella typhimurium, and Aerobacter aerogenes, and Serratia marcescens, it is a bi-functional enzyme with indoleglycerol phosphate synthase as the paired enzyme.[8]
The crystal structure has been characterized for a variety of the above listed microorganisms. The known 2.0 A structure of PRAI from Pyrococcus furiosus shows that tPRAI has a TIM-barrel fold (Fig. 6). PRAI derived from Thermococcus kodakaraensis also expresses a similar TIM-barrel fold structure.[7] The subunits of tPRAI associate via the N-terminal faces of their central beta-barrels. Two long, symmetry-related loops that protrude reciprocally into cavities of the other subunit provide for multiple hydrophobic interactions. Moreover, the side chains of the N-terminal methionines and the C-terminal leucines of both subunits are immobilized in a hydrophobic cluster, and the number of salt bridges is increased in tPRAI. These features appear to be mainly responsible for the high thermostability of tPRAI.[9]
The bi-functional version of this enzyme isolated from E. Coli (Fig. 5) performs two steps within the Tryptophan pathway. Referencing Fig. 7, the N-terminal catalyzes the IGPS reaction (residues ~1–289 purple), and the C-terminal domain performs the PRAI reaction (residues ~158–452 turquoise). Although these domains overlap (orange), the active sites are not overlapping, and studies have shown that mono-functional enzymes composing of these two domains are still able to produce a functional tryptophan bio-synthetic pathway.[10]
The βα loops are responsible for the activity of this enzyme, and the αβ loops are involved in the protein's stability.[8]
More details on the discovery of this enzyme's structure can be found in Willmann's paper.[11]
Active site[7]
Specifically, for phosphoribosyl anthranilate isomerase, TkTrpF, from Thermococcus kodakaraensis. The active site for the Amadori rearrangement, was determined to be Cys8 (acting as the general base) and Asp135 (as the general acid).[12]
Inhibitors
An enzyme inhibitor[13] is molecule that binds to an enzyme that therefore decreases the activity of the protein. The following molecules have been shown to inhibit PRAI activity:
Reduced 1-(2-carboxyphenylamino )-1-deoxy-D-ribulose 5-phosphate [5, 6,8); Indoleglycerol phosphate (8); Indolepropanol phosphate (8); MnCI2 CoCI2 [16); CuS04 (16); More (chemically synthesized N-(5-phospho-betaD-ribosyl)anthranilate contains inhibitors, but not if it is generated by anthranilate phosphoribosyltransferase)
Molecular weight[3]
26300 (Bacillus subtilis, gel filtration)
45000 (Aeromonas formicans, Serratia marinorubra, gel filtration, indole-3-
glycerol-phosphate synthetase/N-5'-phosphoribosylanthranilate isomerase
complex)
46000 (E. coli, sedimentation equilibrium)
47000 (Citrobacter ballerupensis, gel filtration, indole-3-glycerol-phosphate
synthetase/N-5'-phosphoribosylanthranilate isomerase complex)
48000 (Serratia marcescens, Erwinia carotovora, gel filtration, indole-3-glycerol-phosphate synthetase/N-5'-phosphoribosylanthranilate
isomerase complex )
49370 (E. coli, calculated from gene sequence)
53000 (Proteus vulgaris, gel filtration, indole-3-glycerol-phosphate synthetase/
N-5'-phosphoribosylanthranilate isomerase complex)
160000 (Neurospora crassa, gel filtration, component lib of the anthranilate
synthetase complex has N-(5'-phosphoribosyl)anthranilate isomerase and
indole-3-glycerol phosphate synthetase activities)
185000 (Hansenula henricii, gel filtration, indole-3-glycerol-phosphate synthetase/
N-5'-phosphoribosylanthranilate isomerase complex)
Homologous genes
There are homologous genes which produce this enzyme in plant species such as Arabidopsis thaliana and Oryza sativa (Asian Rice). One form of bacterium it is found in Thermotoga maritima.
Phosphoribosylanthranilate isomerase is also found in various forms of fungi such as Kluyveromyces lactis (yeast), Saccharomyces cerevisiae (yeast), and Ashbya gossypii.[14]
A list of genes encoding for PRAI can also be found on KEGG Enzyme database.[15]
References
- ↑ "[46] Chorismate to tryptophan (Escherichia coli)—anthranilate synthetase, PR transferase, PRA isomerase, InGP synthetase, tryptophan synthetase". Metabolism of Amino Acids and Amines Part A. Methods in Enzymology. 17A. 1970. 365–380. doi:10.1016/0076-6879(71)17215-1. ISBN 9780121818746.
- ↑ "TRP1/YDR007W Summary". Saccharomyces genome database. Stanford University. https://www.yeastgenome.org/locus/S000002414.
- ↑ 3.0 3.1 3.2 Schomburg, Dietmar; Stephan, Dörte (1994). Enzyme handbook. Springer-Verlag. ISBN 9783642579424. OCLC 859587801.
- ↑ Lubert Stryer (2019-03-25). Biochemistry. ISBN 9781319114657. OCLC 1052398743.
- ↑ 5.0 5.1 "Phosphoribosyl anthranilate isomerase catalyzes a reversible amadori reaction". Biochemistry 34 (16): 5429–39. April 1995. doi:10.1021/bi00016a014. PMID 7727401.
- ↑ Phosphoribosylanthranilate isomerase and indoleglycerol-phosphate synthase: tryptophan biosynthetic enzymes from Thermotoga maritima. Methods in Enzymology. 331. 2001. pp. 270–80. doi:10.1016/S0076-6879(01)31064-9. ISBN 9780121822323.
- ↑ 7.0 7.1 7.2 Perveen, S.; Rashid, N.; Papageorgiou, A.C. (2016-11-09). Phosphoribosyl anthranilate isomerase from Thermococcus kodakaraensis. doi:10.2210/pdb5lhf/pdb.
- ↑ 8.0 8.1 "Structure and function of mutationally generated monomers of dimeric phosphoribosylanthranilate isomerase from Thermotoga maritima". Structure 8 (3): 265–76. March 2000. doi:10.1016/s0969-2126(00)00106-4. PMID 10745009.
- ↑ "Crystal structure at 2.0 A resolution of phosphoribosyl anthranilate isomerase from the hyperthermophile Thermotoga maritima: possible determinants of protein stability". Biochemistry 36 (20): 6009–16. May 1997. doi:10.1021/bi962718q. PMID 9166771.
- ↑ "Indoleglycerol phosphate synthase-phosphoribosyl anthranilate isomerase: comparison of the bifunctional enzyme from Escherichia coli with engineered monofunctional domains". Biochemistry 34 (16): 5419–28. April 1995. doi:10.1021/bi00016a013. PMID 7727400.
- ↑ PDB: 1PII; "Three-dimensional structure of the bifunctional enzyme phosphoribosylanthranilate isomerase: indoleglycerolphosphate synthase from Escherichia coli refined at 2.0 A resolution". Journal of Molecular Biology 223 (2): 477–507. January 1992. doi:10.1016/0022-2836(92)90665-7. PMID 1738159.
- ↑ Pitt, Charles (2002). Sax, Adolphe (opera). Oxford Music Online. Oxford University Press. doi:10.1093/gmo/9781561592630.article.o006145.
- ↑ "Enzyme→ Inhibitor List: M", Handbook of Enzyme Inhibitors, Wiley-VCH Verlag GmbH, 1999, pp. 894–956, doi:10.1002/9783527618330.ch13, ISBN 9783527618330, https://archive.org/details/handbookofenzyme00helm/page/894
- ↑ "Blast search for phosphoribosylanthranilate isomerase.". HomoloGene Database. National Center for Biotechnology Information.. https://www.ncbi.nlm.nih.gov/homologene/?term=phosphoribosylanthranilate%20isomerase.
- ↑ "KEGG Enzyme". https://www.genome.jp/dbget-bin/www_bget?ec:5.3.1.24.
Further reading
- "The role of the TRP1 gene in yeast tryptophan biosynthesis". The Journal of Biological Chemistry 263 (16): 7868–75. June 1988. doi:10.1016/S0021-9258(18)68578-3. PMID 3286643.
 |