Biology:RNA editing
This article is missing information about pseudouracil. (December 2020) |
| Part of a series on |
| Genetics |
|---|
 |
| Key components |
| History and topics |
| Research |
| Personalized medicine |
| Personalized medicine |
RNA editing (also RNA modification) is a molecular process through which some cells can make discrete changes to specific nucleotide sequences within an RNA molecule after it has been generated by RNA polymerase. It occurs in all living organisms and is one of the most evolutionarily conserved properties of RNAs.[1][2][3] RNA editing may include the insertion, deletion, and base substitution of nucleotides within the RNA molecule. RNA editing is relatively rare, with common forms of RNA processing (e.g. splicing, 5'-capping, and 3'-polyadenylation) not usually considered as editing. It can affect the activity, localization as well as stability of RNAs, and has been linked with human diseases.[1][2][3][4]
RNA editing has been observed in some tRNA, rRNA, mRNA, or miRNA molecules of eukaryotes and their viruses, archaea, and prokaryotes.[5] RNA editing occurs in the cell nucleus, as well as within mitochondria and plastids. In vertebrates, editing is rare and usually consists of a small number of changes to the sequence of the affected molecules. In other organisms, such as squids,[6] extensive editing (pan-editing) can occur; in some cases the majority of nucleotides in an mRNA sequence may result from editing. More than 160 types of RNA modifications have been described so far.[7]
RNA-editing processes show great molecular diversity, and some appear to be evolutionarily recent acquisitions that arose independently. The diversity of RNA editing phenomena includes nucleobase modifications such as cytidine (C) to uridine (U) and adenosine (A) to inosine (I) deaminations, as well as non-template nucleotide additions and insertions. RNA editing in mRNAs effectively alters the amino acid sequence of the encoded protein so that it differs from that predicted by the genomic DNA sequence.[8]

Detection of RNA editing
Next generation sequencing
To identify diverse post-transcriptional modifications of RNA molecules and determine the transcriptome-wide landscape of RNA modifications by means of next generation RNA sequencing, recently many studies have developed conventional[9] or specialised sequencing methods.[1][2][3] Examples of specialised methods are MeRIP-seq,[10] m6A-seq,[11] PA-m5C-seq [12], methylation-iCLIP,[13] m6A-CLIP,[14] Pseudo-seq,[15] Ψ-seq,[16] CeU-seq,[17] Aza-IP[18] and RiboMeth-seq[19]). Many of these methods are based on specific capture of the RNA species containing the specific modification, for example through antibody binding coupled with sequencing of the captured reads. After the sequencing these reads are mapped against the whole transcriptome to see where they originate from.[20] Generally with this kind of approach it is possible to see the location of the modifications together with possible identification of some consensus sequences that might help identification and mapping further on. One example of the specialize methods is PA-m5C-seq. This method was further developed from PA-m6A-seq method to identify m5C modifications on mRNA instead of the original target N6-methyladenosine. The easy switch between different modifications as target is made possible with a simple change of the capturing antibody form m6A specific to m5C specific.[12] Application of these methods have identified various modifications (e.g. pseudouridine, m6A, m5C, 2′-O-Me) within coding genes and non-coding genes (e.g. tRNA, lncRNAs, microRNAs) at single nucleotide or very high resolution.[4]
Mass spectrometry
Mass spectrometry is a way to quantify RNA modifications.[21] More often than not, modifications cause an increase in mass for a given nucleoside. This gives a characteristic readout for the nucleoside and the modified counterpart.[21] Moreover, mass spectrometry allows the investigation of modification dynamics by labelling RNA molecules with stable (non-radioactive) heavy isotopes in vivo. Due to the defined mass increase of heavy isotope labeled nucleosides they can be distinguished from their respective unlabelled isotopomeres by mass spectrometry. This method, called NAIL-MS (nucleic acid isotope labelling coupled mass spectrometry), enables a variety of approaches to investigate RNA modification dynamics.[22][23][24]
Types of RNA
Messenger RNA modification
Recently, functional experiments have revealed many novel functional roles of RNA modifications. Most of the RNA modifications are found on transfer-RNA and ribosomal-RNA, but also eukaryotic mRNA has been shown to be modified with multiple different modifications. 17 naturally occurring modifications on mRNA have been identified, from which the N6-methyladenosine is the most abundant and studied.[25] mRNA modifications are linked to many functions in the cell. They ensure the correct maturation and function of the mRNA, but also at the same time act as part of cell's immune system.[26] Certain modifications like 2'O-methylated nucleotides has been associated with cells ability to distinguish own mRNA from foreign RNA.[27] For example, m6A has been predicted to affect protein translation and localization,[1][2][3] mRNA stability,[28] alternative polyA choice [14] and stem cell pluripotency.[29] Pseudouridylation of nonsense codons suppresses translation termination both in vitro and in vivo, suggesting that RNA modification may provide a new way to expand the genetic code.[30] 5-methylcytosine on the other hand has been associated with mRNA transport from the nucleus to the cytoplasm and enhancement of translation. These functions of m5C are not fully known and proven but one strong argument towards these functions in the cell is the observed localization of m5C to translation initiation site.[31] Importantly, many modification enzymes are dysregulated and genetically mutated in many disease types.[1] For example, genetic mutations in pseudouridine synthases cause mitochondrial myopathy, sideroblastic anemia (MLASA) [32] and dyskeratosis congenital.[33]
Compared to the modifications identified from other RNA species like tRNA and rRNA, the amount of identified modifications on mRNA is very small. One of the biggest reasons why mRNA modifications are not so well known is missing research techniques. In addition to the lack of identified modifications, the knowledge of associated proteins is also behind other RNA species. Modifications are results of specific enzyme interactions with the RNA molecule.[25] Considering mRNA modifications most of the known related enzymes are the writer enzymes that add the modification on the mRNA. The additional groups of enzymes readers and erasers are for most of the modifications either poorly known of not known at all.[34] For these reasons there has been during the past decade huge interest in studying these modifications and their function.[20]
Transfer RNA modifications
Transfer RNA or tRNA is the most abundantly modified type of RNA.[35] Modifications in tRNA play crucial roles in maintaining translation efficiency through supporting structure, anticodon-codon interactions, and interactions with enzymes.[36]
Anticodon modifications are important for proper decoding of mRNA. Since the genetic code is degenerate, anticodon modifications are necessary to properly decode mRNA. Particularly, the wobble position of the anticodon determines how the codons are read. For example, in eukaryotes an adenosine at position 34 of the anticodon can be converted to inosine. Inosine is a modification that is able to base-pair with cytosine, adenine, and uridine.[37]
Another commonly modified base in tRNA is the position adjacent to the anticodon. Position 37 is often hypermodified with bulky chemical modifications. These modifications prevent frameshifting and increase anticodon-codon binding stability through stacking interactions.[37]
Ribosomal RNA modification
Ribosomal RNA (rRNA) is essential to the makeup of ribosomes and peptide transfer during translation processes.[38] Ribosomal RNA modifications are made throughout ribosome synthesis, and often occur during and/or after translation. Modifications primarily play a role in the structure of the rRNA in order to protect translational efficiency.[38] Chemical modification in rRNA consists of methylation of ribose sugars, isomerization of uridines, and methylation and acetylation of individual bases.[39]
Methylation
Methylation of rRNA upholds structural rigidity by blocking base pair stacking and surrounds the 2'-OH group to block hydrolysis. It occurs at specific parts of eukaryotic rRNA. The template for methylation consists of 10-21 nucleotides.[38] 2'-O-methylation of the ribose sugar is one of the most common rRNA modifications.[40] Methylation is primarily introduced by small nucleolar RNA's, referred to as snoRNPs. There are two classes of snoRNPs that target methylation sites, and they are referred to box C/D and box H/ACA.[39][40] One type of methylation, 2′-O-methylation, contributes to helical stabilization.[38]
Isomerization
The isomerization of uridine to pseudouridine is the second most common rRNA modification. These pseudouridines are also introduced by the same classes of snoRNPs that participate in methylation. Pseudouridine synthases are the major participating enzymes in the reaction.[41] The H/ACA box snoRNPs introduce guide sequences that are about 14-15 nucleotides long.[39] Pseudouridylation is triggered in numerous places of rRNAs at once to preserve the thermal stability of RNA.[39] Pseudouridine allows for increased hydrogen bonding and alters translation in rRNA and tRNA.[40][41] It alters translation by increasing the affinity of the ribosome subunit to specific mRNAs.[38]
Base Editing:
Base editing is the third major class of rRNA modification, specifically in eukaryotes. There are 8 categories of base edits that can occur at the gap between the small and large ribosomal subunits.[38] RNA methyltransferases are the enzymes that introduce base methylation.[38] Acetyltransferases are the enzymes responsible for acetylation of cytosine in rRNA. Base methylation plays a role in translation. These base modifications all work in conjunction with the two other main classes of modification to contribute to RNA structural stability. An example of this occurs in N7-methylation, which increases the nucleotide's charge to increase ionic interactions of proteins attaching to the RNA before translation.
Editing by insertion or deletion

RNA editing through the addition and deletion of uracil has been found in kinetoplasts from the mitochondria of Trypanosoma brucei.[42] Because this may involve a large fraction of the sites in a gene, it is sometimes called "pan-editing" to distinguish it from topical editing of one or a few sites.
Pan-editing starts with the base-pairing of the unedited primary transcript with a guide RNA (gRNA), which contains complementary sequences to the regions around the insertion/deletion points. The newly formed double-stranded region is then enveloped by an editosome, a large multi-protein complex that catalyzes the editing.[43][44] The editosome opens the transcript at the first mismatched nucleotide and starts inserting uridines. The inserted uridines will base-pair with the guide RNA, and insertion will continue as long as A or G is present in the guide RNA and will stop when a C or U is encountered.[45][46] The inserted nucleotides cause a frameshift, and result in a translated protein that differs from its gene.
The mechanism of the editosome involves an endonucleolytic cut at the mismatch point between the guide RNA and the unedited transcript. The next step is catalyzed by one of the enzymes in the complex, a terminal U-transferase, which adds Us from UTP at the 3' end of the mRNA.[47] The opened ends are held in place by other proteins in the complex. Another enzyme, a U-specific exoribonuclease, removes the unpaired Us. After editing has made mRNA complementary to gRNA, an RNA ligase rejoins the ends of the edited mRNA transcript.[48][49] As a consequence, the editosome can edit only in a 3' to 5' direction along the primary RNA transcript. The complex can act on only a single guide RNA at a time. Therefore, a RNA transcript requiring extensive editing will need more than one guide RNA and editosome complex.
Editing by deamination
C-to-U editing

The editing involves cytidine deaminase that deaminates a cytidine base into a uridine base. An example of C-to-U editing is with the apolipoprotein B gene in humans. Apo B100 is expressed in the liver and apo B48 is expressed in the intestines. In the intestines, the mRNA has a CAA sequence edited to be UAA, a stop codon, thus producing the shorter B48 form. C-to-U editing often occurs in the mitochondrial RNA of flowering plants. Different plants have different degrees of C-to-U editing; for example, eight editing events occur in mitochondria of the moss Funaria hygrometrica, whereas over 1,700 editing events occur in the lycophytes Isoetes engelmanii.[50] C-to-U editing is performed by members of the pentatricopeptide repeat (PPR) protein family. Angiosperms have large PPR families, acting as trans -factors for cis -elements lacking a consensus sequence; Arabidopsis has around 450 members in its PPR family. There have been a number of discoveries of PPR proteins in both plastids and mitochondria.[51]
A-to-I editing
Adenosine-to-inosine (A-to-I) modifications contribute to nearly 90% of all editing events in RNA. The deamination of adenosine is catalyzed by the double-stranded RNA-specific adenosine deaminase (ADAR), which typically acts on pre-mRNAs. The deamination of adenosine to inosine disrupts and destabilizes the dsRNA base pairing, therefore rendering that particular dsRNA less able to produce siRNA, which interferes with the RNAi pathway.
The wobble base pairing causes deaminated RNA to have a unique but different structure, which may be related to the inhibition of the initiation step of RNA translation. Studies have shown that I-RNA (RNA with many repeats of the I-U base pair) recruits methylases that are involved in the formation of heterochromatin and that this chemical modification heavily interferes with miRNA target sites.[52] There is active research into the importance of A-to-I modifications and their purpose in the novel concept of epitranscriptomics, in which modifications are made to RNA that alter their function.[53][54] A long established consequence of A-to-I in mRNA is the interpretation of I as a G, therefore leading to functional A-to-G substitution, e.g. in the interpretation of the genetic code by ribosomes. Newer studies, however, have weakened this correlation by showing that inosines can also be decoded by the ribosome (although in a lesser extent) as adenosines or uracils. Furthermore, it was shown that I's lead to the stalling of ribosomes on the I-rich mRNA.[55]
The development of high-throughput sequencing in recent years has allowed for the development of extensive databases for different modifications and edits of RNA. RADAR (Rigorously Annotated Database of A-to-I RNA editing) was developed in 2013 to catalog the vast variety of A-to-I sites and tissue-specific levels present in humans, mice, and flies. The addition of novel sites and overall edits to the database are ongoing.[56] The level of editing for specific editing sites, e.g. in the filamin A transcript, is tissue-specific.[57] The efficiency of mRNA-splicing is a major factor controlling the level of A-to-I RNA editing.[58][59] Interestingly, ADAR1 and ADAR2 also affect alternative splicing via both A-to-I editing ability and dsRNA binding ability.[60][61]
Alternative mRNA editing
Alternative U-to-C mRNA editing was first reported in WT1 (Wilms Tumor-1) transcripts,[62] and non-classic G-A mRNA changes were first observed in HNRNPK (heterogeneous nuclear ribonucleoprotein K) transcripts in both malignant and normal colorectal samples.[63] The latter changes were also later seen alongside non-classic U-to-C alterations in brain cell TPH2 (tryptophan hydroxylase 2) transcripts.[64] Although the reverse amination might be the simplest explanation for U-to-C changes, transamination and transglycosylation mechanisms have been proposed for plant U-to-C editing events in mitochondrial transcripts.[65] A recent study reported novel G-to-A mRNA changes in WT1 transcripts at two hotspots, proposing the APOBEC3A (apolipoprotein B mRNA editing enzyme, catalytic polypeptide 3A) as the enzyme implicated in this class of alternative mRNA editing.[66] It was also shown that alternative mRNA changes were associated with canonical WT1 splicing variants, indicating their functional significance.
RNA editing in plant mitochondria and plastids
It has been shown in previous studies that the only types of RNA editing seen in the plants' mitochondria and plastids are conversion of C-to-U and U-to-C (very rare).[67][68][69][70][71][72][73][74][75][76][77][78][79] RNA-editing sites are found mainly in the coding regions of mRNA, introns, and other non-translated regions.[69] In fact, RNA editing can restore the functionality of tRNA molecules.[71][72] The editing sites are found primarily upstream of mitochondrial or plastid RNAs. While the specific positions for C to U RNA editing events have been fairly well studied in both the mitochondrion and plastid,[80] the identity and organization of all proteins comprising the editosome have yet to be established. Members of the expansive PPR protein family have been shown to function as trans-acting factors for RNA sequence recognition.[81] Specific members of the MORF (Multiple Organellar RNA editing Factor) family are also required for proper editing at several sites. As some of these MORF proteins have been shown to interact with members of the PPR family, it is possible MORF proteins are components of the editosome complex.[82] An enzyme responsible for the trans- or deamination of the RNA transcript remains elusive, though it has been proposed that the PPR proteins may serve this function as well.
RNA editing is essential for the normal functioning of the plant's translation and respiration activity. Editing can restore the essential base-pairing sequences of tRNAs, restoring functionality.[83] It has also been linked to the production of RNA-edited proteins that are incorporated into the polypeptide complexes of the respiration pathway. Therefore, it is highly probable that polypeptides synthesized from unedited RNAs would not function properly and hinder the activity of both mitochondria and plastids.
C-to-U RNA editing can create start and stop codons, but it cannot destroy existing start and stop codons. A cryptic start codon is created when the codon ACG is edited to be AUG.
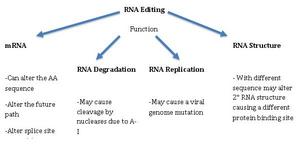
RNA editing in viruses
Viruses (i.e., measles, mumps, or parainfluenza), especially viruses that have an RNA genome, have been shown to have evolved to utilize RNA modifications in many ways when taking over the host cell. Viruses are known to utilize the RNA modifications in different parts of their infection cycle from immune evasion to protein translation enhancement.[27] RNA editing is used for stability and generation of protein variants.[84][85] Viral RNAs are transcribed by a virus-encoded RNA-dependent RNA polymerase, which is prone to pausing and "stuttering" at certain nucleotide combinations. In addition, up to several hundred non-templated A's are added by the polymerase at the 3' end of nascent mRNA.[86] These As help stabilize the mRNA. Furthermore, the pausing and stuttering of the RNA polymerase allows the incorporation of one or two Gs or As upstream of the translational codon.[86] The addition of the non-templated nucleotides shifts the reading frame, which generates a different protein.
Additionally, the RNA modifications are shown to have both positive and negative effects on the replication and translation efficiency depending on the virus. For example, Courtney et al.[12] showed that an RNA modification called 5-methylcytosine is added to the viral mRNA in infected host cells in order to enhance the protein translation of HIV-1 virus. The inhibition of the m5C modification on viral mRNA results in significant reduction in viral protein translation, but interestingly it has no effect on the expression of viral mRNAs in the cell. On the other hand, Lichinchi et al.[87] showed that the N6-methyladenosine modification on ZIKV mRNA inhibits the viral replication.
Origin and evolution of RNA editing
The RNA-editing system seen in the animal may have evolved from mononucleotide deaminases, which have led to larger gene families that include the apobec-1 and adar genes. These genes share close identity with the bacterial deaminases involved in nucleotide metabolism. The adenosine deaminase of E. coli cannot deaminate a nucleoside in the RNA; the enzyme's reaction pocket is too small for the RNA strand to bind to. However, this active site is widened by amino acid changes in the corresponding human analog genes, APOBEC1 and ADAR, allowing deamination.[88][89] The gRNA-mediated pan-editing in trypanosome mitochondria, involving templated insertion of U residues, is an entirely different biochemical reaction. The enzymes involved have been shown in other studies to be recruited and adapted from different sources.[43][90] But the specificity of nucleotide insertion via the interaction between the gRNA and mRNA is similar to the tRNA editing processes in the animal and Acanthamoeba mitochondria.[91] Eukaryotic ribose methylation of rRNAs by guide RNA molecules is a similar form of modification.[92]
Thus, RNA editing evolved more than once. Several adaptive rationales for editing have been suggested.[93] Editing is often described as a mechanism of correction or repair to compensate for defects in gene sequences. However, in the case of gRNA-mediated editing, this explanation does not seem possible because if a defect happens first, there is no way to generate an error-free gRNA-encoding region, which presumably arises by duplication of the original gene region. A more plausible alternative for the evolutionary origins of this system is through constructive neutral evolution, where the order of steps is reversed, with the gratuitous capacity for editing preceding the "defect".[94]
Therapeutic mRNA editing
Directing edits to correct mutated sequences was first proposed and demonstrated in 1995.[95] This initial work used synthetic RNA antisense oligonucleotides complementary to a pre-mature stop codon mutation in a dystrophin sequence to activate A-to-I editing of the stop codon to a read through codon in a model xenopus cell system.[95] While this also led to nearby inadvertent A-to-I transitions, A to I (read as G) transitions can correct all three stop codons, but cannot create a stop codon. Therefore, the changes led >25% correction of the targeted stop codon with read through to a downstream luciferase reporter sequence. Follow on work by Rosenthal achieved editing of mutated mRNA sequence in mammalian cell culture by directing an oligonucleotide linked to a cytidine deaminase to correct a mutated cystic fibrosis sequence.[96] More recently, CRISPR-Cas13 fused to deaminases has been employed to direct mRNA editing.[97]
In 2022, therapeutic RNA editing for Cas7-11 was reported.[98][99] It enables sufficiently targeted cuts and an early version of it was used for in vitro editing in 2021.[100]
Comparison to DNA editing
Unlike DNA editing, which is permanent, the effects of RNA editing − including potential off-target mutations in RNA − are transient and are not inherited. RNA editing is therefore considered to be less risky. Furthermore, it may only require a guide RNA by using the ADAR protein already found in humans and many other eukaryotes' cells instead of needing to introduce a foreign protein into the body.[101]
See also
- DNA editing
- Epigenome editing
- NcRNA therapy
- RNA timestamp
References
- ↑ 1.0 1.1 1.2 1.3 1.4 "The pivotal regulatory landscape of RNA modifications". Annual Review of Genomics and Human Genetics 15: 127–150. 2013. doi:10.1146/annurev-genom-090413-025405. PMID 24898039.
- ↑ 2.0 2.1 2.2 2.3 "Mapping recently identified nucleotide variants in the genome and transcriptome". Nature Biotechnology 30 (11): 1107–1116. November 2012. doi:10.1038/nbt.2398. PMID 23138310.
- ↑ 3.0 3.1 3.2 3.3 "The dynamic epitranscriptome: N6-methyladenosine and gene expression control". Nature Reviews. Molecular Cell Biology 15 (5): 313–326. May 2014. doi:10.1038/nrm3785. PMID 24713629.
- ↑ 4.0 4.1 "RMBase: a resource for decoding the landscape of RNA modifications from high-throughput sequencing data". Nucleic Acids Research 44 (D1): D259–D265. January 2016. doi:10.1093/nar/gkv1036. PMID 26464443.
- ↑ "A-to-I and C-to-U editing within transfer RNAs". Biochemistry. Biokhimiia 76 (8): 932–937. August 2011. doi:10.1134/S0006297911080098. PMID 22022967.
- ↑ "New genetic editing powers discovered in squid" (in en-us). phys.org. https://phys.org/news/2020-03-genetic-powers-squid.html.
- ↑ "MODOMICS: a database of RNA modification pathways. 2017 update". Nucleic Acids Research 46 (D1): D303–D307. January 2018. doi:10.1093/nar/gkx1030. PMID 29106616.
- ↑ "RNA editing". FEMS Microbiology Reviews 23 (3): 297–316. June 1999. doi:10.1111/j.1574-6976.1999.tb00401.x. PMID 10371035.
- ↑ "Accurate mapping of tRNA reads". Bioinformatics (Oxford, England) 34 (7): 1116–1124. April 2018. doi:10.1093/bioinformatics/btx756. PMID 29228294.
- ↑ "Comprehensive analysis of mRNA methylation reveals enrichment in 3' UTRs and near stop codons". Cell 149 (7): 1635–1646. June 2012. doi:10.1016/j.cell.2012.05.003. PMID 22608085.
- ↑ "Topology of the human and mouse m6A RNA methylomes revealed by m6A-seq". Nature 485 (7397): 201–206. April 2012. doi:10.1038/nature11112. PMID 22575960. Bibcode: 2012Natur.485..201D.
- ↑ 12.0 12.1 12.2 "Epitranscriptomic Addition of m5C to HIV-1 Transcripts Regulates Viral Gene Expression". Cell Host & Microbe 26 (2): 217–227.e6. August 2019. doi:10.1016/j.chom.2019.07.005. PMID 31415754.
- ↑ "NSun2-mediated cytosine-5 methylation of vault noncoding RNA determines its processing into regulatory small RNAs". Cell Reports 4 (2): 255–261. July 2013. doi:10.1016/j.celrep.2013.06.029. PMID 23871666.
- ↑ 14.0 14.1 "A majority of m6A residues are in the last exons, allowing the potential for 3' UTR regulation". Genes & Development 29 (19): 2037–2053. October 2015. doi:10.1101/gad.269415.115. PMID 26404942.
- ↑ "Pseudouridine profiling reveals regulated mRNA pseudouridylation in yeast and human cells". Nature 515 (7525): 143–146. November 2014. doi:10.1038/nature13802. PMID 25192136. Bibcode: 2014Natur.515..143C.
- ↑ "Transcriptome-wide mapping reveals widespread dynamic-regulated pseudouridylation of ncRNA and mRNA". Cell 159 (1): 148–162. September 2014. doi:10.1016/j.cell.2014.08.028. PMID 25219674.
- ↑ "Chemical pulldown reveals dynamic pseudouridylation of the mammalian transcriptome". Nature Chemical Biology 11 (8): 592–597. August 2015. doi:10.1038/nchembio.1836. PMID 26075521.
- ↑ "Identification of direct targets and modified bases of RNA cytosine methyltransferases". Nature Biotechnology 31 (5): 458–464. May 2013. doi:10.1038/nbt.2566. PMID 23604283.
- ↑ "Profiling of ribose methylations in RNA by high-throughput sequencing". Angewandte Chemie 54 (2): 451–455. January 2015. doi:10.1002/anie.201408362. PMID 25417815. Bibcode: 2015ACIE...54..451B.
- ↑ 20.0 20.1 "Mapping and editing of nucleic acid modifications". Computational and Structural Biotechnology Journal 18: 661–667. 2020. doi:10.1016/j.csbj.2020.03.010. PMID 32257049.
- ↑ 21.0 21.1 "Mass spectrometry of modified RNAs: recent developments". The Analyst 141 (1): 16–23. January 2016. doi:10.1039/C5AN01797A. PMID 26501195. Bibcode: 2016Ana...141...16W.
- ↑ "Observing the fate of tRNA and its modifications by nucleic acid isotope labeling mass spectrometry: NAIL-MS". RNA Biology 14 (9): 1260–1268. September 2017. doi:10.1080/15476286.2017.1325063. PMID 28488916.
- ↑ "NAIL-MS in E. coli Determines the Source and Fate of Methylation in tRNA". ChemBioChem 19 (24): 2575–2583. December 2018. doi:10.1002/cbic.201800525. PMID 30328661.
- ↑ "Surpassing limits of static RNA modification analysis with dynamic NAIL-MS". Methods 156: 91–101. March 2019. doi:10.1016/j.ymeth.2018.10.025. PMID 30395967.
- ↑ 25.0 25.1 "Naturally occurring modified ribonucleosides". Wiley Interdisciplinary Reviews. RNA 11 (5). September 2020. doi:10.1002/wrna.1595. PMID 32301288.
- ↑ "The chemical diversity of RNA modifications". The Biochemical Journal 476 (8): 1227–1245. April 2019. doi:10.1042/BCJ20180445. PMID 31028151.
- ↑ 27.0 27.1 "Epitranscriptomic regulation of viral replication". Biochimica et Biophysica Acta (BBA) - Gene Regulatory Mechanisms 1860 (4): 460–471. April 2017. doi:10.1016/j.bbagrm.2017.02.002. PMID 28219769. https://repositorio.uchile.cl/handle/2250/167060.
- ↑ "N6-methyladenosine-dependent regulation of messenger RNA stability". Nature 505 (7481): 117–120. January 2014. doi:10.1038/nature12730. PMID 24284625. Bibcode: 2014Natur.505..117W.
- ↑ "Stem cells. m6A mRNA methylation facilitates resolution of naïve pluripotency toward differentiation". Science 347 (6225): 1002–1006. February 2015. doi:10.1126/science.1261417. PMID 25569111.
- ↑ "Converting nonsense codons into sense codons by targeted pseudouridylation". Nature 474 (7351): 395–398. June 2011. doi:10.1038/nature10165. PMID 21677757.
- ↑ "5-methylcytosine promotes mRNA export - NSUN2 as the methyltransferase and ALYREF as an m5C reader". Cell Research 27 (5): 606–625. May 2017. doi:10.1038/cr.2017.55. PMID 28418038.
- ↑ "Missense mutation in pseudouridine synthase 1 (PUS1) causes mitochondrial myopathy and sideroblastic anemia (MLASA)". American Journal of Human Genetics 74 (6): 1303–1308. June 2004. doi:10.1086/421530. PMID 15108122.
- ↑ "X-linked dyskeratosis congenita is caused by mutations in a highly conserved gene with putative nucleolar functions". Nature Genetics 19 (1): 32–38. May 1998. doi:10.1038/ng0598-32. PMID 9590285.
- ↑ "Messenger RNA modifications: Form, distribution, and function". Science 352 (6292): 1408–1412. June 2016. doi:10.1126/science.aad8711. PMID 27313037. Bibcode: 2016Sci...352.1408G.
- ↑ "Emerging roles of tRNA in adaptive translation, signalling dynamics and disease". Nature Reviews. Genetics 16 (2): 98–112. February 2015. doi:10.1038/nrg3861. PMID 25534324.
- ↑ "tRNA Modifications: Impact on Structure and Thermal Adaptation". Biomolecules 7 (2): 35. April 2017. doi:10.3390/biom7020035. PMID 28375166.
- ↑ 37.0 37.1 "tRNA's wobble decoding of the genome: 40 years of modification". Journal of Molecular Biology 366 (1): 1–13. February 2007. doi:10.1016/j.jmb.2006.11.046. PMID 17187822.
- ↑ 38.0 38.1 38.2 38.3 38.4 38.5 38.6 "Tuning the ribosome: The influence of rRNA modification on eukaryotic ribosome biogenesis and function". RNA Biology 14 (9): 1138–1152. September 2017. doi:10.1080/15476286.2016.1259781. PMID 27911188.
- ↑ 39.0 39.1 39.2 39.3 Sharma, Sunny; Entian, Karl-Dieter (2022), Entian, Karl-Dieter, ed., "Chemical Modifications of Ribosomal RNA" (in en), Ribosome Biogenesis: Methods and Protocols, Methods in Molecular Biology (New York, NY: Springer US) 2533: pp. 149–166, doi:10.1007/978-1-0716-2501-9_9, ISBN 978-1-0716-2501-9, PMID 35796987
- ↑ 40.0 40.1 40.2 Borchardt, Erin K.; Martinez, Nicole M.; Gilbert, Wendy V. (2020-11-23). "Regulation and Function of RNA Pseudouridylation in Human Cells". Annual Review of Genetics 54: 309–336. doi:10.1146/annurev-genet-112618-043830. ISSN 0066-4197. PMID 32870730.
- ↑ 41.0 41.1 Borchardt, Erin K.; Martinez, Nicole M.; Gilbert, Wendy V. (2020-11-23). "Regulation and Function of RNA Pseudouridylation in Human Cells" (in en). Annual Review of Genetics 54 (1): 309–336. doi:10.1146/annurev-genet-112618-043830. ISSN 0066-4197. PMID 32870730.
- ↑ "RNA editing in trypanosomes". European Journal of Biochemistry 221 (1): 9–23. April 1994. doi:10.1111/j.1432-1033.1994.tb18710.x. PMID 7513284.
- ↑ 43.0 43.1 "Mechanism and evolution of RNA editing in kinetoplastida". Biochimica et Biophysica Acta (BBA) - Gene Structure and Expression 1307 (1): 39–54. June 1996. doi:10.1016/0167-4781(96)00021-8. PMID 8652667.
- ↑ "The mechanism of U insertion/deletion RNA editing in kinetoplastid mitochondria". Nucleic Acids Research 25 (19): 3751–3759. October 1997. doi:10.1093/nar/25.19.3751. PMID 9380494.
- ↑ "A model for RNA editing in kinetoplastid mitochondria: "guide" RNA molecules transcribed from maxicircle DNA provide the edited information". Cell 60 (2): 189–198. January 1990. doi:10.1016/0092-8674(90)90735-W. PMID 1688737.
- ↑ "RNA editing: getting U into RNA". Trends in Biochemical Sciences 22 (5): 162–166. May 1997. doi:10.1016/S0968-0004(97)01041-4. PMID 9175474.
- ↑ "Sense from nonsense: RNA editing in mitochondria of kinetoplastid protozoa and slime molds". Cell 81 (6): 837–840. June 1995. doi:10.1016/0092-8674(95)90003-9. PMID 7781060.
- ↑ "RNA editing in mitochondrial mRNA of trypanosomatids". Trends in Biochemical Sciences 16 (2): 68–72. February 1991. doi:10.1016/0968-0004(91)90027-S. PMID 1713359.
- ↑ "Mitochondrial mRNA editing in kinetoplastid protozoa". Modification and Editing of RNA. Washington, DC.: ASM Press. 1998. pp. 377–394.
- ↑ "RNA editing in plant mitochondria—connecting RNA target sequences and acting proteins". Mitochondrion. Plant Mitochondria in Mitochondrion 19 (Pt B): 191–197. November 2014. doi:10.1016/j.mito.2014.04.005. PMID 24732437.
- ↑ "RNA editing in plants: Machinery and flexibility of site recognition". Biochimica et Biophysica Acta (BBA) - Bioenergetics. SI: Chloroplast Biogenesis 1847 (9): 779–785. September 2015. doi:10.1016/j.bbabio.2014.12.010. PMID 25585161.
- ↑ "Functions and regulation of RNA editing by ADAR deaminases". Annual Review of Biochemistry 79 (1): 321–349. 2010. doi:10.1146/annurev-biochem-060208-105251. PMID 20192758.
- ↑ "The dynamic epitranscriptome: A to I editing modulates genetic information". Chromosoma 125 (1): 51–63. March 2016. doi:10.1007/s00412-015-0526-9. PMID 26148686.
- ↑ "Rapid and dynamic transcriptome regulation by RNA editing and RNA modifications". The Journal of Cell Biology 213 (1): 15–22. April 2016. doi:10.1083/jcb.201511041. PMID 27044895.
- ↑ "Inosine induces context-dependent recoding and translational stalling". Nucleic Acids Research 47 (1): 3–14. January 2019. doi:10.1093/nar/gky1163. PMID 30462291.
- ↑ "RADAR: a rigorously annotated database of A-to-I RNA editing". Nucleic Acids Research 42 (Database issue): D109–D113. January 2014. doi:10.1093/nar/gkt996. PMID 24163250.
- ↑ "Spatio-temporal profiling of Filamin A RNA-editing reveals ADAR preferences and high editing levels outside neuronal tissues". RNA Biology 10 (10): 1611–1617. October 2013. doi:10.4161/rna.26216. PMID 24025532.
- ↑ "Adenosine to Inosine editing frequency controlled by splicing efficiency". Nucleic Acids Research 44 (13): 6398–6408. July 2016. doi:10.1093/nar/gkw325. PMID 27112566.
- ↑ "A high resolution A-to-I editing map in the mouse identifies editing events controlled by pre-mRNA splicing". Genome Research 29 (9): 1453–1463. September 2019. doi:10.1101/gr.242636.118. PMID 31427386.
- ↑ "ADAR-deficiency perturbs the global splicing landscape in mouse tissues". Genome Research 30 (8): 1107–1118. August 2020. doi:10.1101/gr.256933.119. PMID 32727871.
- ↑ "Cis- and trans-regulations of pre-mRNA splicing by RNA editing enzymes influence cancer development". Nature Communications 11 (1). February 2020. doi:10.1038/s41467-020-14621-5. PMID 32034135. Bibcode: 2020NatCo..11..799T.
- ↑ "RNA editing in the Wilms' tumor susceptibility gene, WT1". Genes & Development 8 (6): 720–731. March 1994. doi:10.1101/gad.8.6.720. PMID 7926762.
- ↑ "Editing of hnRNP K protein mRNA in colorectal adenocarcinoma and surrounding mucosa". British Journal of Cancer 94 (4): 586–592. February 2006. doi:10.1038/sj.bjc.6602938. PMID 16404425.
- ↑ "Alternative splicing and extensive RNA editing of human TPH2 transcripts". PLOS ONE 5 (1). January 2010. doi:10.1371/journal.pone.0008956. PMID 20126463. Bibcode: 2010PLoSO...5.8956G.
- ↑ "RNA editing in plant organelles. Why make it easy?". Biochemistry. Biokhimiia 76 (8): 924–931. August 2011. doi:10.1134/S0006297911080086. PMID 22022966.
- ↑ "APOBEC3A is implicated in a novel class of G-to-A mRNA editing in WT1 transcripts". PLOS ONE 10 (3). 2015. doi:10.1371/journal.pone.0120089. PMID 25807502. Bibcode: 2015PLoSO..1020089N.
- ↑ "RNA editing in plant mitochondria". Nature 341 (6243): 662–666. October 1989. doi:10.1038/341662a0. PMID 2552326. Bibcode: 1989Natur.341..662C.
- ↑ "RNA editing in wheat mitochondria results in the conservation of protein sequences". Nature 341 (6243): 660–662. October 1989. doi:10.1038/341660a0. PMID 2552325. Bibcode: 1989Natur.341..660G.
- ↑ 69.0 69.1 "RNA editing in plant mitochondria". Science 246 (4937): 1632–1634. December 1989. doi:10.1126/science.2480644. PMID 2480644. Bibcode: 1989Sci...246.1632H.
- ↑ "Editing of a chloroplast mRNA by creation of an initiation codon". Nature 353 (6340): 178–180. September 1991. doi:10.1038/353178a0. PMID 1653905. Bibcode: 1991Natur.353..178H.
- ↑ 71.0 71.1 "RNA editing gives a new meaning to the genetic information in mitochondria and chloroplasts". Plant Molecular Biology 21 (6): 1163–1170. March 1993. doi:10.1007/BF00023611. PMID 8490134. Bibcode: 1993PMolB..21.1163P.
- ↑ 72.0 72.1 "Regenerating good sense: RNA editing and trans splicing in plant mitochondria". Trends in Genetics 8 (9): 322–328. September 1992. doi:10.1016/0168-9525(92)90265-6. PMID 1365399.
- ↑ "RNA editing in plant organelles". RNA Editing (Benne, R., Ed.), Ellis Harwood, New York.. 1993.
- ↑ "RNA editing in bryophytes and a molecular phylogeny of land plants". The EMBO Journal 15 (6): 1403–1411. March 1996. doi:10.1002/j.1460-2075.1996.tb00482.x. PMID 8635473.
- ↑ "Occurrence of plastid RNA editing in all major lineages of land plants". Proceedings of the National Academy of Sciences of the United States of America 94 (12): 6285–6290. June 1997. doi:10.1073/pnas.94.12.6285. PMID 9177209. Bibcode: 1997PNAS...94.6285F.
- ↑ "Editing and import: strategies for providing plant mitochondria with a complete set of functional transfer RNAs". Biochimie 78 (6): 518–529. 1996. doi:10.1016/0300-9084(96)84758-4. PMID 8915541.
- ↑ "Identification of critical nucleotide positions for plastid RNA editing site recognition". RNA 3 (10): 1194–1200. October 1997. PMID 9326494.
- ↑ "RNA editing in plant mitochondria and chloroplasts". FASEB Journal 7 (1): 64–71. January 1993. doi:10.1096/fasebj.7.1.8422976. PMID 8422976.
- ↑ "Preface". Modification and Editing of RNA. Washington, DC: ASM Press. 1998. pp. 307–323.
- ↑ "RNA editing in plants and its evolution". Annual Review of Genetics 47: 335–352. 2013. doi:10.1146/annurev-genet-111212-133519. PMID 24274753.
- ↑ "Pentatricopeptide repeat proteins in plants". Annual Review of Plant Biology 65 (1): 415–442. 2014. doi:10.1146/annurev-arplant-050213-040159. PMID 24471833. Bibcode: 2014AnRPB..65..415B.
- ↑ "Comprehensive high-resolution analysis of the role of an Arabidopsis gene family in RNA editing". PLOS Genetics 9 (6). June 2013. doi:10.1371/journal.pgen.1003584. PMID 23818871.
- ↑ "Editing of tRNA". Modification and Editing of RNA. Washington, DC: ASM Press. 1998. pp. 289–306.
- ↑ "The Sendai virus P gene expresses both an essential protein and an inhibitor of RNA synthesis by shuffling modules via mRNA editing". The EMBO Journal 10 (10): 3079–3085. October 1991. doi:10.1002/j.1460-2075.1991.tb07860.x. PMID 1655410.
- ↑ "Editing on the genomic RNA of human hepatitis delta virus". Journal of Virology 66 (8): 4693–4697. August 1992. doi:10.1128/jvi.66.8.4693-4697.1992. PMID 1629949.
- ↑ 86.0 86.1 "Chapter 23: Cotranscriptional Paramyxovirus mRNA Editing: a Contradiction in Terms?". Modification and Editing of RNA. Washington, DC: ASM Press. 1998. pp. 413–420.
- ↑ "Dynamics of Human and Viral RNA Methylation during Zika Virus Infection". Cell Host & Microbe 20 (5): 666–673. November 2016. doi:10.1016/j.chom.2016.10.002. PMID 27773536.
- ↑ "Nucleoside deaminases for cytidine and adenosine: comparisons with deaminases acting on RNA". Modification and Editing of RNA. Washington, DC: ASM Press. 1998. pp. 363–376.
- ↑ "On the evolution of RNA editing". Trends in Genetics 9 (8): 265–268. August 1993. doi:10.1016/0168-9525(93)90011-6. PMID 8379005.
- ↑ "Predicted editing of additional transfer RNAs in Acanthamoeba castellanii mitochondria". Nucleic Acids Research 21 (18): 4402. September 1993. doi:10.1093/nar/21.18.4402. PMID 8415006.
- ↑ "Small nucleolar RNAs guide the ribose methylations of eukaryotic rRNAs". Modification and Editing of RNA. Washington, DC: ASM Press. 1998. pp. 255–272.
- ↑ "Does constructive neutral evolution play an important role in the origin of cellular complexity? Making sense of the origins and uses of biological complexity". BioEssays 33 (5): 344–349. May 2011. doi:10.1002/bies.201100010. PMID 21381061.
- ↑ "On the possibility of constructive neutral evolution". Journal of Molecular Evolution 49 (2): 169–181. August 1999. doi:10.1007/PL00006540. PMID 10441669. Bibcode: 1999JMolE..49..169S.
- ↑ 95.0 95.1 "Toward the therapeutic editing of mutated RNA sequences". Proceedings of the National Academy of Sciences of the United States of America 92 (18): 8298–8302. August 1995. doi:10.1073/pnas.92.18.8298. PMID 7545300. Bibcode: 1995PNAS...92.8298W.
- ↑ "Correction of mutations within the cystic fibrosis transmembrane conductance regulator by site-directed RNA editing". Proceedings of the National Academy of Sciences of the United States of America 110 (45): 18285–18290. November 2013. doi:10.1073/pnas.1306243110. PMID 24108353. Bibcode: 2013PNAS..11018285M.
- ↑ "RNA editing with CRISPR-Cas13". Science 358 (6366): 1019–1027. November 2017. doi:10.1126/science.aaq0180. PMID 29070703. Bibcode: 2017Sci...358.1019C.
- ↑ "Neuroscientists expand CRISPR toolkit with new, compact Cas7-11 enzyme" (in en). Massachusetts Institute of Technology. https://phys.org/news/2022-05-neuroscientists-crispr-toolkit-compact-cas7-.html.
- ↑ "Structure and engineering of the type III-E CRISPR-Cas7-11 effector complex". Cell 185 (13): 2324–2337.e16. June 2022. doi:10.1016/j.cell.2022.05.003. PMID 35643083.
- ↑ "Programmable RNA targeting with the single-protein CRISPR effector Cas7-11". Nature 597 (7878): 720–725. September 2021. doi:10.1038/s41586-021-03886-5. PMID 34489594. Bibcode: 2021Natur.597..720O.
- ↑ "Watch out, CRISPR. The RNA editing race is on" (in en). Chemical & Engineering News 97 (12). 25 March 2019. https://cen.acs.org/business/start-ups/WatchCRISPR-RNA-editing-race/97/i12. Retrieved 30 September 2020.
 |
