Biology:RNF227
RING Finger Protein 227, also known as RNF227 and LINC02581, is a protein which in humans is encoded by the RNF227 gene.[1] According to DNA microarray data, it is found in at least 15 tissues.[1] [citation needed]
Gene
In humans, the RNF227 gene is found on chromosome 17 p13.1. Its mRNA sequence is 2850 base pairs in length and includes 2 exons. The coding sequence is from base pairs 95 to 2835.[2]
Protein
The RNF227 protein is 190 amino acids in length, seen in the table below.[3]

thumb|Predicted tertiary structure.[5]
1 |
MQLLVRVPSL PERGELDCNI CYRPFNLGCR APRRLPGTAR ARCGHTICTA CLRELAARGD |
61 |
GGGAAARVVR LRRVVTCPFC RAPSQLPRGG LTEMALDSDL WSRLEEKARA KCERDEAGNP |
121 |
AKESSDADGE AEEEGESEKG AGPRSAGWRA LRRLWDRVLG PARRWRRPLP SNVLYCAEIK |
181 |
DIGHLTRCTL |
Predicted properties
Using tools at Expasy, the predicted molecular weight of the protein sequence is 20,875 kilodaltons[3] with an isoelectric point of 9.23.[6] The Statistical Analysis of Protein Sequences tool detected two repetitive structures: CRAPRRLP from positions 29 to 36 and CRAPSQLP from positions 80 to 87.[7]
Zinc finger domain
RING Finger Protein 227 has a zinc finger domain from position 18 to 81 , which is highly conserved throughout many eukaryotic organisms.[8]
Secondary structure
The secondary structure was predicted by the I-TASSER server and shows 7 alpha helices, 4 beta strands, and 12 coils.[4]
Tertiary structure
The tertiary structure was predicted by the I-TASSER with a confidence score of -3.42, which is typically in the range from -5 to 2.[4]
Gene level regulation
RNA-seq was performed of tissue samples from 95 human individuals representing 27 different tissues in order to determine tissue-specificity of all protein-coding genes. The highest expression can be seen in the skin, with an expression value of 22 ± 4.5 Reads per Kilobase of transcript, per Million mapped reads (RPKM). Transcription profiling was done by high throughput sequencing of individual and mixtures of 16 human tissues RNA to show the highest expression in the testes. Additionally, the lowest expression is seen in the liver. RNA sequencing was conducted of the total RNA from 20 human tissues which showed high expression in the brain, both in the cerebellum and fetal tissues. 35 human fetal samples from 6 tissues (3 – 7 replicates per tissue) collected between 10- and 20-weeks gestational time were sequence using Illumina TruSeq Stranded Total RNA. This shows very high expression in the intestine after 11 weeks and the kidney after 10 weeks.[1]
Three experiments were found that show what conditions RNF227 rises and falls. A study conducted on T cell-driven IL-22 amplification of Il-1beta-driven inflammation in human adipose tissue shows how there is higher expression of RNF227 in obese non-diabetic patients.[9] An analysis of non-invasive NeuN cells and invasive NeuT cells treated with interstitial fluid flow resulted in higher expression of RNF227 in the NeuN cell line in both the static and flow protocols. This gives insight into the molecular pathways activated by interstitial fluid flow in ERBB2-positive breast cancer cells. [10] The last experiment showed how the effect of Rho kinase inhibition on long-term keratinocyte proliferation is rapid and conditional and resulted in higher expression in the control agent as compared to the Y-27632 agent. [11]
Transcript level regulation
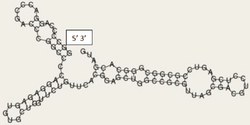

The diagram to the right depicts the stem-loop formation of the 5' untranslated region of RNF227.[12] The BED4.02, ZFX.01, and ZIC3.03 transcription factors are seen with RNF227, which is notable because they are all associated with zinc finger domains.[13] Translation is initiated at the AUG start codon, as seen in the conceptual translation.
Protein level regulation
The Motif Scan tool at MyHits predicted casein kinase II phosphorylation sites (from positions 9 to 12, 102 to 105, and 125 to 128), N-myristylation sites (from positions 37 to 42 and 61 to 66), and protein kinase c phosphorylation sites (from positions 38 to 40 and 137 to 139).[14]
Additionally, PSORT II predicted a 69.6% chance for the protein sequence to be found in the nucleus of a cell.[15]
Homology



RING Finger Protein 227 has no paralogs. It does, however, have numerous orthologs extending throughout eukaryotes. The following table presents a selection of orthologs found using searches in BLAST[19] and BLAT.[20] This is not meant to be a comprehensive list, rather a small sample that shows the diversity of species in which orthologs are found.
| Genus and Species | Common Name | Taxonomic Group | Date of Divergence (Million Years Ago) | Accession Number | Sequence Length (amino acids) | Sequence Identity | Sequence Similarity |
|---|---|---|---|---|---|---|---|
| Homo sapiens | Human | Primates | 0 | NP_001345628.1 | 190 | 100% | 100% |
| Neotoma lepida | Desert Woodrat | Rodentia | 90 | OBS67541.1 | 164 | 67.9% | 73.7% |
| Microtus ochrogaster | Prairie vole | Rodentia | 90 | XP_026636787.1 | 214 | 67.4% | 74.4% |
| Dipodomys ordii | Ord's Kangaroo Rat | Rodentia | 90 | XP_012868576.1 | 158 | 64.8% | 68.9% |
| Balaenoptera acutorostrata scammoni | Minke Whale | Artiodactyla | 90 | XP_028024073.1 | 166 | 65.3% | 71.1% |
| Vulpes vulpes | Red Fox | Carnivora | 96 | XP_025861213.1 | 160 | 62.8% | 72.3% |
| Vicugna pacos | Alpaca | Artiodactyla | 96 | XP_006218277.1 | 156 | 24.8% | 30.4% |
| Vombatus ursinus | Common Wombat | Diprotondontia | 159 | XP_027712916.1 | 180 | 62.0% | 74.5% |
| Sarcophilus harrisii | Tasmanian Devil | Dasyuromorphia | 159 | XP_023358488.2 | 172 | 58.8% | 66.2% |
| Gallus gallus | Chicken | Galliformes | 312 | XP_001234238.1 | 168 | 25.9% | 36.1% |
| Geotrypetes seraphini | Gaboon Caecilian | Gymnophiona | 351.8 | XP_033780950.1 | 150 | 37.2% | 46.9% |
| Rhinatrema bivittatum | Two-lined Caecilian | Gymnophiona | 351.8 | XP_029437562.1 | 152 | 35.7% | 48.0% |
| Microcaecilia unicolor | Cayenne Caecilian | Gymnophiona | 351.8 | XP_030043188.1 | 148 | 34.4% | 46.9% |
| Xenopus tropicalis | Western Clawed Frog | Anura | 351.8 | XP_031750786.1 | 178 | 19.1% | 43.4% |
| Scleropages formosus | Asian Arowana | Osteoglossiformes | 435 | XP_029113159.1 | 165 | 30.6% | 44.9% |
| Astyanax mexicanus | Mexican Tetra | Characiformes | 435 | XP_007231481.2 | 161 | 28.0% | 37.4% |
| Paramormyrops kinsleyae | Old Calabar Mormyrid | Osteoglossiformes | 435 | XP_023674393.1 | 165 | 26.6% | 32.3% |
| Lepisosteus oculatus | Spotted Gar | Lepisosteiformes | 435 | XP_006640609.2 | 172 | 24.6% | 33.0% |
| Salmo trutta | Brown Trout | Saloniformes | 435 | XP_029622555.1 | 190 | 24.2% | 29.6% |
| Danio rerio | Zebrafish | Cypriniformes | 435 | NP_001121828.1 | 187 | 23.6% | 35.6% |
References
- ↑ 1.0 1.1 1.2 "RNF227 ring finger protein 227 [Homo sapiens (human) - Gene - NCBI"]. https://www.ncbi.nlm.nih.gov/gene/284023.
- ↑ Homo sapiens ring finger protein 227 (RNF227), transcript variant 1, mRNA - Nucleotide - NCBI. 9 June 2022. http://www.ncbi.nlm.nih.gov/nuccore/NM_001358699.2.
- ↑ 3.0 3.1 "RING finger protein 227 [Homo sapiens - Protein - NCBI"]. https://www.ncbi.nlm.nih.gov/protein/NP_001345628.1.
- ↑ 4.0 4.1 4.2 "I-TASSER results". https://zhanglab.ccmb.med.umich.edu/I-TASSER/output/S583327/.
- ↑ "RNF227 ring finger protein 227 [Homo sapiens (human) - Gene - NCBI"]. https://www.ncbi.nlm.nih.gov/gene/284023.
- ↑ "Compute pI/MW". https://web.expasy.org/cgi-bin/compute_pi/pi_tool.[yes|permanent dead link|dead link}}]
- ↑ "SAPS Results". https://www.ebi.ac.uk/Tools/services/web/toolresult.ebi?jobId=saps-I20201219-200406-0679-86942162-p1m.
- ↑ "RNF227 - RING finger protein 227 - Homo sapiens (Human) - RNF227 gene & protein". https://www.uniprot.org/uniprot/A6NIN4.
- ↑ "Type 2 diabetic obese patients: visceral adipose tissue CD14+ cells". https://www.ncbi.nlm.nih.gov/geo/tools/profileGraph.cgi?ID=GDS5167:ILMN_1739325.
- ↑ "Interstitial fluid flow effect on noninvasive and invasive ERBB2-positive breast cancer cells". https://www.ncbi.nlm.nih.gov/geo/tools/profileGraph.cgi?ID=GDS5800:ILMN_1739325.
- ↑ "Rho kinase inhibition effect on epidermal keratinocyte in vitro". https://www.ncbi.nlm.nih.gov/geo/tools/profileGraph.cgi?ID=GDS5431:ILMN_1739325.
- ↑ "RNAfold web server". http://rna.tbi.univie.ac.at//cgi-bin/RNAWebSuite/RNAfold.cgi?PAGE=3&ID=WSCplEQQao%5D.
- ↑ "Genomatix: Retrieve and analyze promoters: Query Input". https://www.genomatix.de/cgi-bin/c2p/c2p.pl?s=686e67c37f2213fa866d8f0dfcac089f;C2P_SELECTION=1.
- ↑ "Motif Scan" (in en). https://myhits.sib.swiss/cgi-bin/motif_scan#GRAPHIC.
- ↑ "PSORT II Prediction". https://psort.hgc.jp/cgi-bin/runpsort.pl.[yes|permanent dead link|dead link}}]
- ↑ "Clustal Omega < Multiple Sequence Alignment < EMBL-EBI". https://www.ebi.ac.uk/Tools/msa/clustalo/.
- ↑ "Clustal Omega < Multiple Sequence Alignment < EMBL-EBI". https://www.ebi.ac.uk/Tools/msa/clustalo/.
- ↑ "Clustal Omega < Multiple Sequence Alignment < EMBL-EBI". https://www.ebi.ac.uk/Tools/msa/clustalo/.
- ↑ "BLAST: Basic Local Alignment Search Tool". https://blast.ncbi.nlm.nih.gov/Blast.cgi.
- ↑ "Human BLAT Search". https://genome.ucsc.edu/cgi-bin/hgBlat.
 |
