Biology:Teichoic acid
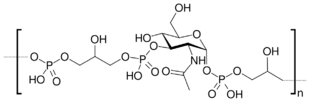
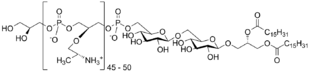
Teichoic acids (cf. Greek τεῖχος, teīkhos, "wall", to be specific a fortification wall, as opposed to τοῖχος, toīkhos, a regular wall)[1] are bacterial copolymers [2] of glycerol phosphate or ribitol phosphate and carbohydrates linked via phosphodiester bonds.
Teichoic acids are found within the cell wall of most Gram-positive bacteria such as species in the genera Staphylococcus, Streptococcus, Bacillus, Clostridium, Corynebacterium, and Listeria, and appear to extend to the surface of the peptidoglycan layer. They can be covalently linked to N-acetylmuramic acid or a terminal D-alanine in the tetrapeptide crosslinkage between N-acetylmuramic acid units of the peptidoglycan layer, or they can be anchored in the cytoplasmic membrane with a lipid anchor. Teichoic acid's chemical signal is CH17P4O29NOH.
Teichoic acids that are anchored to the lipid membrane are referred to as lipoteichoic acids (LTAs), whereas teichoic acids that are covalently bound to peptidoglycan are referred to as wall teichoic acids (WTA).[3]
Structure
The most common structure of Wall teichoic acids are a ManNAc(β1→4)GlcNAc disaccharide with one to three glycerol phosphates attached to the C4 hydroxyl of the ManNAc residue followed by a long chain of glycerol- or ribitol phosphate repeats.[3] Variations come in the long chain tail, which generally include sugar subunits being attached to the sides or the body of the repeats. Four types of WTA repeats have been named, as of 2013.[4]
Lipoteichoic acids follow a similar pattern of putting most variation in the repeats, although the set of enzymes used are different, at least in the case of Type I LTA. The repeats are anchored onto the membrane via a (di)glucosyl-diacylglycerol (Glc(2)DAG) anchor. Type IV LTA from Streptococcus pneumoniae represents a special case where both types intersect: after the tail is synthesized with an undecaprenyl phosphate (C55-P) intermediate "head", different TagU/LCP (LytR-CpsA-Psr) family enzymes either attaches it to the wall to form a WTA or to the GlcDAG anchor.[5]
Function
The main function of teichoic acids is to provide flexibility to the cell-wall by attracting cations such as calcium and potassium. Teichoic acids can be substituted with D-alanine ester residues,[6] or D-glucosamine,[7] giving the molecule zwitterionic properties.[8] These zwitterionic teichoic acids are suspected ligands for toll-like receptors 2 and 4. Teichoic acids also assist in regulation of cell growth by limiting the ability of autolysins to break the β(1-4) bond between the N-acetyl glucosamine and the N-acetylmuramic acid.
Lipoteichoic acids may also act as receptor molecules for some Gram-positive bacteriophage; however, this has not yet been conclusively supported.[9] It is an acidic polymer and contributes negative charge to the cell wall.
Biosynthesis
WTA and Type IV LTA
Enzymes involved in the biosynthesis of WTAs have been named: TarO, TarA, TarB, TarF, TarK, and TarL. Their roles are:[3]
- TarO (O34753, EC 2.7.8.33) starts off the process by connecting GlcNAc to a biphospho-undecaprenyl (bactoprenyl) in the inner membrane.
- TarA (P27620, EC 2.4.1.187) connects a ManNAc to the UDP-GlcNac formed by TarO via a β-(1,4) linkage.
- TarB (P27621, EC 2.7.8.44) connects a single glycerol-3-phosphate to the C4 hydroxyl of ManNAc.
- TarF (P13485, EC 2.7.8.12) connects more glycerol-3-phosphate units to the glycerol tail. In Tag-producing bacteria, this is the final step (a long glycerol tail). Otherwise it only adds one unit.
- TarK (Q8RKJ1, EC 2.7.8.46) connects the initial ribitol-5-phosphate unit. It is necessary in Bacillus subtilis W23 for Tar production, but S. aureus has both functions in the same TarL/K enzyme.
- TarL (Q8RKJ2, EC 2.7.8.47) constructs the long ribitol-5-phosphate tail.
Following the synthesis, the ATP-binding cassette transporters (teichoic-acid-transporting ATPase) TarGH (P42953, P42954) flip the cytoplasmic complex to the external surface of the inner membrane. The redundant TagTUV enzymes link this product to the cell wall.[4] The enzymes TarI (Q8RKI9) and TarJ (Q8RKJ0) are responsible for producing the substrates that lead to the polymer tail. Many of these proteins are located in a conserved gene cluster.[3]
Later (2013) studies have identified a few more enzymes that attach unique sugars to the WTA repeat units. A set of enzymes and transporters named DltABCE that adds alanines to both wall and lipo-teichoic acids were found.[4]
Note that the set of genes are named "Tag" (teichoic acid glycerol) instead of "Tar" (teichoic acid ribitol) in B. subtilis 168, which lacks the TarK/TarL enzymes. TarB/F/L/K all bear some similarities to each other, and belong to the same family (InterPro: IPR007554).[3] Due to the role of B. subtilis as the main model strain, some linked UniProt entries are in fact the "Tag" ortholog as they are better annotated. The "similarity search" may be used to access the genes in the Tar-producing B. substilis W23 (BACPZ).
As an antibiotic drug target
This was proposed in 2004.[3] A further review in 2013 has given more specific parts of the pathways to inhibit given newer knowledge.[4]
See also
- Lipoteichoic acid – a major constituent of the cell wall of gram-positive bacteria
- Sir James Baddiley
References
- ↑ τεῖχος. Liddell, Henry George; Scott, Robert; A Greek–English Lexicon at the Perseus Project
- ↑ Teichoic+acids at the US National Library of Medicine Medical Subject Headings (MeSH)
- ↑ 3.0 3.1 3.2 3.3 3.4 3.5 "Wall teichoic acid function, biosynthesis, and inhibition". ChemBioChem 11 (1): 35–45. January 2010. doi:10.1002/cbic.200900557. PMID 19899094.
- ↑ 4.0 4.1 4.2 4.3 "Wall teichoic acids of gram-positive bacteria". Annual Review of Microbiology 67 (1): 313–36. 8 September 2013. doi:10.1146/annurev-micro-092412-155620. PMID 24024634.
- ↑ "Lipoteichoic acid synthesis and function in gram-positive bacteria". Annual Review of Microbiology 68 (1): 81–100. 8 September 2014. doi:10.1146/annurev-micro-091213-112949. PMID 24819367.
- ↑ "Immunological properties of teichoic acids". Bacteriological Reviews 37 (2): 215–57. June 1973. doi:10.1128/br.37.2.215-257.1973. PMID 4578758.
- ↑ "Lipoteichoic acid in Streptomyces hygroscopicus: structural model and immunomodulatory activities". PLOS ONE 6 (10): e26316. October 2011. doi:10.1371/journal.pone.0026316. PMID 22028855. Bibcode: 2011PLoSO...626316C.
- ↑ "Conformation of the phosphate D-alanine zwitterion in bacterial teichoic acid from nuclear magnetic resonance spectroscopy". Biochemistry 48 (39): 9242–9. October 2009. doi:10.1021/bi900503k. PMID 19746945.
- ↑ "Molecular interaction between lipoteichoic acids and Lactobacillus delbrueckii phages depends on D-alanyl and alpha-glucose substitution of poly(glycerophosphate) backbones". Journal of Bacteriology 189 (11): 4135–40. June 2007. doi:10.1128/JB.00078-07. PMID 17416656.
External links
- Bioinformatic mappings (see also the EC entries):
- UniProt: WTA KW-0777, pathway:547.789 (poly(glucopyranosyl N-acetylgalactosamine 1-phosphate) teichoic acid biosynthesis), pathway:547.827 (poly(glycerol phosphate) teichoic acid biosynthesis), pathway:547.790 (poly(ribitol phosphate) teichoic acid biosynthesis)
- UniProt: LTA pathway:547.556 (lipoteichoic acid biosynthesis)
- gene ontology: GO:0019350
 |
