Medicine:Medical microbiology

Medical microbiology, the large subset of microbiology that is applied to medicine, is a branch of medical science concerned with the prevention, diagnosis and treatment of infectious diseases. In addition, this field of science studies various clinical applications of microbes for the improvement of health. There are four kinds of microorganisms that cause infectious disease: bacteria, fungi, parasites and viruses, and one type of infectious protein called prion.
A medical microbiologist studies the characteristics of pathogens, their modes of transmission, mechanisms of infection and growth. The academic qualification as a clinical/Medical Microbiologist in a hospital or medical research centre generally requires a Bachelors degree while in some countries a Masters in Microbiology along with Ph.D. in any of the life-sciences (Biochem, Micro, Biotech, Genetics, etc.).[1] Medical microbiologists often serve as consultants for physicians, providing identification of pathogens and suggesting treatment options. Using this information, a treatment can be devised. Other tasks may include the identification of potential health risks to the community or monitoring the evolution of potentially virulent or resistant strains of microbes, educating the community and assisting in the design of health practices. They may also assist in preventing or controlling epidemics and outbreaks of disease. Not all medical microbiologists study microbial pathology; some study common, non-pathogenic species to determine whether their properties can be used to develop antibiotics or other treatment methods.
Epidemiology, the study of the patterns, causes, and effects of health and disease conditions in populations, is an important part of medical microbiology, although the clinical aspect of the field primarily focuses on the presence and growth of microbial infections in individuals, their effects on the human body, and the methods of treating those infections. In this respect the entire field, as an applied science, can be conceptually subdivided into academic and clinical sub-specialties, although in reality there is a fluid continuum between public health microbiology and clinical microbiology, just as the state of the art in clinical laboratories depends on continual improvements in academic medicine and research laboratories.
History


In 1676, Anton van Leeuwenhoek observed bacteria and other microorganisms, using a single-lens microscope of his own design.[3]
In 1796, Edward Jenner developed a method using cowpox to successfully immunize a child against smallpox. The same principles are used for developing vaccines today.[4]
Following on from this, in 1857 Louis Pasteur also designed vaccines against several diseases such as anthrax, fowl cholera and rabies as well as pasteurization for food preservation.[5]
In 1867 Joseph Lister is considered to be the father of antiseptic surgery. By sterilizing the instruments with diluted carbolic acid and using it to clean wounds, post-operative infections were reduced, making surgery safer for patients.[6]
In the years between 1876 and 1884 Robert Koch provided much insight into infectious diseases. He was one of the first scientists to focus on the isolation of bacteria in pure culture. This gave rise to the germ theory, a certain microorganism being responsible for a certain disease. He developed a series of criteria around this that have become known as the Koch's postulates.[7]
A major milestone in medical microbiology is the Gram stain. In 1884 Hans Christian Gram developed the method of staining bacteria to make them more visible and differentiated under a microscope. This technique is widely used today.[8]
In 1910 Paul Ehrlich tested multiple combinations of arsenic based chemicals on infected rabbits with syphilis. Ehrlich then found that arsphenamine was found effective against syphilis spirochetes. The arsphenamines was then made available in 1910, known as Salvarsan.[9]
In 1929 Alexander Fleming developed one of the most commonly used antibiotic substances both at the time and now: penicillin.[10]
In 1939 Gerhard Domagk found Prontosil red protected mice from pathogenic streptococci and staphylococci without toxicity. Domagk received the Nobel Prize in physiology, or medicine, for the discovery of the sulfa drug.[9]
DNA sequencing, a method developed by Walter Gilbert and Frederick Sanger in 1977,[11] caused a rapid change the development of vaccines, medical treatments and diagnostic methods. Some of these include synthetic insulin which was produced in 1979 using recombinant DNA and the first genetically engineered vaccine was created in 1986 for hepatitis B.
In 1995 a team at The Institute for Genomic Research sequenced the first bacterial genome; Haemophilus influenzae.[12] A few months later, the first eukaryotic genome was completed. This would prove invaluable for diagnostic techniques.[13]
In 2007, a team at the Danish food company Danisco, were able to identify the purpose of the CRIPR-Cas systems as adaptive immunity to phages. The system was then quickly found to be able to help in genome editing through its ability to generate double strand breaks. A patient with sickle cell disease was the first person to be treated for a genetic disorder with CRISPR in July 2019.[14]
Commonly treated infectious diseases
Bacterial
- Rotavirus[19]
- Hepatitis C[20]
- Human papillomavirus (HPV)[21]
Causes and transmission of infectious diseases
Infections may be caused by bacteria, viruses, fungi, and parasites. The pathogen that causes the disease may be exogenous (acquired from an external source; environmental, animal or other people, e.g. Influenza) or endogenous (from normal flora e.g. Candidiasis).[27]
The site at which a microbe enters the body is referred to as the portal of entry.[28] These include the respiratory tract, gastrointestinal tract, genitourinary tract, skin, and mucous membranes.[29] The portal of entry for a specific microbe is normally dependent on how it travels from its natural habitat to the host.[28]
There are various ways in which disease can be transmitted between individuals. These include:[28]
- Direct contact - Touching an infected host, including sexual contact
- Indirect contact - Touching a contaminated surface
- Droplet contact - Coughing or sneezing
- Fecal–oral route - Ingesting contaminated food or water sources
- Airborne transmission - Pathogen carrying spores
- Vector transmission - An organism that does not cause disease itself but transmits infection by conveying pathogens from one host to another
- Fomite transmission - An inanimate object or substance capable of carrying infectious germs or parasites
- Environmental - Hospital-acquired infection (Nosocomial infections)
Like other pathogens, viruses use these methods of transmission to enter the body, but viruses differ in that they must also enter into the host's actual cells. Once the virus has gained access to the host's cells, the virus' genetic material (RNA or DNA) must be introduced to the cell. Replication between viruses is greatly varied and depends on the type of genes involved in them. Most DNA viruses assemble in the nucleus while most RNA viruses develop solely in cytoplasm.[30][31]
The mechanisms for infection, proliferation, and persistence of a virus in cells of the host are crucial for its survival. For example, some diseases such as measles employ a strategy whereby it must spread to a series of hosts. In these forms of viral infection, the illness is often treated by the body's own immune response, and therefore the virus is required to disperse to new hosts before it is destroyed by immunological resistance or host death.[32] In contrast, some infectious agents such as the Feline leukemia virus, are able to withstand immune responses and are capable of achieving long-term residence within an individual host, whilst also retaining the ability to spread into successive hosts.[33]
Diagnostic tests
Identification of an infectious agent for a minor illness can be as simple as clinical presentation; such as gastrointestinal disease and skin infections. In order to make an educated estimate as to which microbe could be causing the disease, epidemiological factors need to be considered; such as the patient's likelihood of exposure to the suspected organism and the presence and prevalence of a microbial strain in a community.
Diagnosis of infectious disease is nearly always initiated by consulting the patient's medical history and conducting a physical examination. More detailed identification techniques involve microbial culture, microscopy, biochemical tests and genotyping. Other less common techniques (such as X-rays, CAT scans, PET scans or NMR) are used to produce images of internal abnormalities resulting from the growth of an infectious agent.
Microbial culture
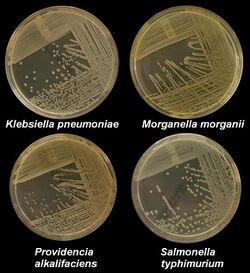
Microbiological culture is the primary method used for isolating infectious disease for study in the laboratory. Tissue or fluid samples are tested for the presence of a specific pathogen, which is determined by growth in a selective or differential medium.
The 3 main types of media used for testing are:[34]
- Solid culture: A solid surface is created using a mixture of nutrients, salts and agar. A single microbe on an agar plate can then grow into colonies (clones where cells are identical to each other) containing thousands of cells. These are primarily used to culture bacteria and fungi.
- Liquid culture: Cells are grown inside a liquid media. Microbial growth is determined by the time taken for the liquid to form a colloidal suspension. This technique is used for diagnosing parasites and detecting mycobacteria.[35]
- Cell culture: Human or animal cell cultures are infected with the microbe of interest. These cultures are then observed to determine the effect the microbe has on the cells. This technique is used for identifying viruses.
Microscopy
Culture techniques will often use a microscopic examination to help in the identification of the microbe. Instruments such as compound light microscopes can be used to assess critical aspects of the organism. This can be performed immediately after the sample is taken from the patient and is used in conjunction with biochemical staining techniques, allowing for resolution of cellular features. Electron microscopes and fluorescence microscopes are also used for observing microbes in greater detail for research.[36] The two main types of electron microscopy are scanning electron microscopy and transmission electron microscopy. Transmission electron microscopy passes electrons through a thin cross-section of the cell of interest, and it then redirects the electrons onto a fluorescent screen. This method is useful for looking at the inside of cells, and the structures within, especially cell walls and membranes. Scanning electron microscopy reads the electrons that are reflected off the surface of the cells. A 3-dimensional image is then made which shows the size and exterior structure of the cells. Both techniques help give more detailed information about the structure of microbes. This makes it useful in many medical fields, such as diagnostics and biopsies of many body parts, hygiene, and virology. They provide critical information about the structure of pathogens, which allow physicians to treat them with more knowledge.[37]
Biochemical tests
Fast and relatively simple biochemical tests can be used to identify infectious agents. For bacterial identification, the use of metabolic or enzymatic characteristics are common due to their ability to ferment carbohydrates in patterns characteristic of their genus and species. Acids, alcohols and gases are usually detected in these tests when bacteria are grown in selective liquid or solid media, as mentioned above. In order to perform these tests en masse, automated machines are used. These machines perform multiple biochemical tests simultaneously, using cards with several wells containing different dehydrated chemicals. The microbe of interest will react with each chemical in a specific way, aiding in its identification.
Serological methods are highly sensitive, specific and often extremely rapid laboratory tests used to identify different types of microorganisms. The tests are based upon the ability of an antibody to bind specifically to an antigen. The antigen (usually a protein or carbohydrate made by an infectious agent) is bound by the antibody, allowing this type of test to be used for organisms other than bacteria. This binding then sets off a chain of events that can be easily and definitively observed, depending on the test. More complex serological techniques are known as immunoassays. Using a similar basis as described above, immunoassays can detect or measure antigens from either infectious agents or the proteins generated by an infected host in response to the infection.[34]
Polymerase chain reaction
Polymerase chain reaction (PCR) assays are the most commonly used molecular technique to detect and study microbes.[38] As compared to other methods, sequencing and analysis is definitive, reliable, accurate, and fast.[39] Today, quantitative PCR is the primary technique used, as this method provides faster data compared to a standard PCR assay. For instance, traditional PCR techniques require the use of gel electrophoresis to visualize amplified DNA molecules after the reaction has finished. quantitative PCR does not require this, as the detection system uses fluorescence and probes to detect the DNA molecules as they are being amplified.[40] In addition to this, quantitative PCR also removes the risk of contamination that can occur during standard PCR procedures (carrying over PCR product into subsequent PCRs).[38] Another advantage of using PCR to detect and study microbes is that the DNA sequences of newly discovered infectious microbes or strains can be compared to those already listed in databases, which in turn helps to increase understanding of which organism is causing the infectious disease and thus what possible methods of treatment could be used.[39] This technique is the current standard for detecting viral infections such as AIDS and hepatitis.
Treatments
Once an infection has been diagnosed and identified, suitable treatment options must be assessed by the physician and consulting medical microbiologists. Some infections can be dealt with by the body's own immune system, but more serious infections are treated with antimicrobial drugs. Bacterial infections are treated with antibacterials (often called antibiotics) whereas fungal and viral infections are treated with antifungals and antivirals respectively. A broad class of drugs known as antiparasitics are used to treat parasitic diseases.
Medical microbiologists often make treatment recommendations to the patient's physician based on the strain of microbe and its antibiotic resistances, the site of infection, the potential toxicity of antimicrobial drugs and any drug allergies the patient has.
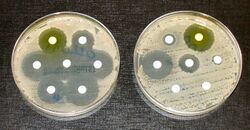
In addition to drugs being specific to a certain kind of organism (bacteria, fungi, etc.), some drugs are specific to a certain genus or species of organism, and will not work on other organisms. Because of this specificity, medical microbiologists must consider the effectiveness of certain antimicrobial drugs when making recommendations. Additionally, strains of an organism may be resistant to a certain drug or class of drug, even when it is typically effective against the species. These strains, termed resistant strains, present a serious public health concern of growing importance to the medical industry as the spread of antibiotic resistance worsens. Antimicrobial resistance is an increasingly problematic issue that leads to millions of deaths every year.[41]
Whilst drug resistance typically involves microbes chemically inactivating an antimicrobial drug or a cell mechanically stopping the uptake of a drug, another form of drug resistance can arise from the formation of biofilms. Some bacteria are able to form biofilms by adhering to surfaces on implanted devices such as catheters and prostheses and creating an extracellular matrix for other cells to adhere to.[42] This provides them with a stable environment from which the bacteria can disperse and infect other parts of the host. Additionally, the extracellular matrix and dense outer layer of bacterial cells can protect the inner bacteria cells from antimicrobial drugs.[43]
Phage therapy is a technique that was discovered before antibiotics, but fell to the wayside as antibiotics became predominate. It is now being considered as a potential solution to increasing antimicrobial resistance. Bacteriophages, viruses that only infect bacteria, can specifically target the bacteria of interest and inject their genome. This process makes the bacteria halt its own production to make more phages, and this continues until the bacteria lyses itself and releases the phages into the surrounding environment. Phage therapy does not kill microbiota since it is specific, and it can help those with antibiotic allergies. Some drawbacks are that it is a time-intensive process since the specific bacterium needs to be identified. It also does not currently have the body of research supporting its effects and safety that antibiotics do. Bacteria can also eventually become resistant, through systems like CRISPR/Cas9 system. Many clinical trials have been promising though, showing that it could potentially help with the antimicrobial resistance problem. It can also be used in conjunction with antibiotics for a cumulative effect.[44]
Medical microbiology is not only about diagnosing and treating disease, it also involves the study of beneficial microbes. Microbes have been shown to be helpful in combating infectious disease and promoting health. Treatments can be developed from microbes, as demonstrated by Alexander Fleming's discovery of penicillin as well as the development of new antibiotics from the bacterial genus Streptomyces among many others.[45] Not only are microorganisms a source of antibiotics but some may also act as probiotics to provide health benefits to the host, such as providing better gastrointestinal health or inhibiting pathogens.[46]
References
- ↑ Thomson, R. B.; Wilson, M. L.; Weinstein, M. P. (2010). "The Clinical Microbiology Laboratory Director in the United States Hospital Setting". Journal of Clinical Microbiology 48 (10): 3465–3469. doi:10.1128/JCM.01575-10. PMID 20739497.
- ↑ Tan, S. Y.; Berman, E. (2008). "Robert Koch (1843-1910): father of microbiology and Nobel laureate". Singapore Medical Journal 49 (11): 854–855. PMID 19037548. https://pubmed.ncbi.nlm.nih.gov/19037548.
- ↑ Frank N. Egerton (2006). "A History of the Ecological Sciences, Part 19: Leeuwenhoek's Microscopic Natural History". Bulletin of the Ecological Society of America 87: 47–58. doi:10.1890/0012-9623(2006)87[47:AHOTES2.0.CO;2].
- ↑ Riedel, Stefan (2005). "Edward Jenner and the history of smallpox and vaccination". Proceedings (Baylor University. Medical Center) 18 (1): 21–25. doi:10.1080/08998280.2005.11928028. ISSN 0899-8280. PMID 16200144.
- ↑ Madigan M, ed (2006). Brock Biology of Microorganisms (13th ed.). Pearson Education. p. 1096. ISBN 978-0-321-73551-5.
- ↑ Michaleas, Spyros N.; Laios, Konstantinos; Charalabopoulos, Alexandros; Samonis, George; Karamanou, Marianna; Michaleas, Spyridon; Laios, Konstantinos; Charalabopoulos, Alexandros et al. (2022-12-21). "Joseph Lister (1827-1912): A Pioneer of Antiseptic Surgery" (in en). Cureus 14 (12): e32777. doi:10.7759/cureus.32777. ISSN 2168-8184. PMID 36686094.
- ↑ Brock TD (1999). Robert Koch: a life in medicine and bacteriology. Washington DC: American Society of Microbiology Press. ISBN 978-1-55581-143-3.
- ↑ Coico, Richard (2006). "Gram Staining" (in en). Current Protocols in Microbiology Appendix 3 (1): Appendix 3C. doi:10.1002/9780471729259.mca03cs00. ISSN 1934-8525. PMID 18770544. https://currentprotocols.onlinelibrary.wiley.com/doi/10.1002/9780471729259.mca03cs00.
- ↑ 9.0 9.1 Willey, Joanne; Sandman, Kathleen; Wood, Dorothy (2020). Prescott's Microbiology. New York: McGraw-Hill Education. pp. 188. ISBN 978-1-260-21188-7.
- ↑ Gaynes, Robert (2017). "The Discovery of Penicillin—New Insights After More Than 75 Years of Clinical Use" (in en-us). Emerging Infectious Diseases 23 (5): 849–853. doi:10.3201/eid2305.161556. PMC 5403050. https://wwwnc.cdc.gov/eid/article/23/5/16-1556_article.
- ↑ Sanger F, Nicklen S, Coulson AR (1977) DNA sequencing with chain-terminating inhibitors" Proceedings of the National Academy of Sciences 74:5463-5467.
- ↑ Fleischmann R, Adams M, White O, Clayton R, Kirkness E, Kerlavage A, Bult C, Tomb J, Dougherty B, Merrick J, al. e (1995) Whole-genome random sequencing and assembly of Haemophilus influenzae Rd" Science 269:496-512.
- ↑ Prescott LM, Harley JP, Klein DA (2005) Microbiology: McGraw-Hill Higher Education.
- ↑ (Stein 2019) (Barrangou & Horvath 2017)
- ↑ Shaikh N; Leonard E; Martin JM (September 2010). "Prevalence of streptococcal pharyngitis and streptococcal carriage in children: a meta-analysis". Pediatrics 126 (3): 557–564. doi:10.1542/peds.2009-2648. PMID 20696723. http://pediatrics.aappublications.org/content/126/3/e557.abstract.
- ↑ Vos T (December 2012). "Years lived with disability (YLDs) for 1160 sequelae of 289 diseases and injuries 1990–2010: a systematic analysis for the Global Burden of Disease Study 2010". Lancet 380 (9859): 2163–96. doi:10.1016/S0140-6736(12)61729-2. PMID 23245607.
- ↑ "Typhoid Fever". World Health Organization. https://www.who.int/vaccine_research/diseases/diarrhoeal/en/index7.html.
- ↑ 18.0 18.1 "World Health Statistics 2012". World Health Organization. https://www.who.int/gho/publications/world_health_statistics/2012/en/index.html.
- ↑ Dennehy PH (2012). "Rotavirus infection: an update on management and prevention". Advances in Pediatrics 59 (1): 47–74. doi:10.1016/j.yapd.2012.04.002. PMID 22789574.
- ↑ "Hepatitis C". World Health Organization. https://www.who.int/mediacentre/factsheets/fs164/en/index.html.
- ↑ Dunne EF; Unger ER; Sternberg, M (February 2007). "Prevalence of HPV infection among females in the United States". Journal of the American Medical Association 297 (8): 813–9. doi:10.1001/jama.297.8.813. PMID 17327523.
- ↑ Kappus KD; Lundgren RG Jr.; Juranek DD; Roberts JM et al. (June 1994). "Intestinal parasitism in the United States: update on a continuing problem". The American Journal of Tropical Medicine and Hygiene 50 (6): 705–13. doi:10.4269/ajtmh.1994.50.705. PMID 8024063.
- ↑ "Toxoplasmosis". Centers for Disease Control and Prevention. https://www.cdc.gov/parasites/toxoplasmosis/.
- ↑ "Candidiasis". Centers for Disease Control and Prevention. https://www.cdc.gov/fungal/candidiasis/genital/statistics.html.
- ↑ "Histoplasmosis". Centers for Disease Control and Prevention. https://www.cdc.gov/fungal/histoplasmosis/statistics.html.
- ↑ Nenoff, P.; Krüger, C.; Mayser, P. (2015-06-01). "[Cutaneous Malassezia infections and Malassezia associated dermatoses: An update"]. Der Hautarzt; Zeitschrift für Dermatologie, Venerologie, und Verwandte Gebiete 66 (6): 465–484; quiz 485–486. doi:10.1007/s00105-015-3631-z. ISSN 1432-1173. PMID 25968082. https://pubmed.ncbi.nlm.nih.gov/25968082/.
- ↑ Washington, JA (1996). "10 Principles of Diagnosis". in Baron, S. Medical Microbiology (4th ed.). University of Texas Medical Branch at Galveston. ISBN 978-0-9631172-1-2. https://www.ncbi.nlm.nih.gov/books/NBK8014/.
- ↑ 28.0 28.1 28.2 Siebeling, RJ (1998). "Chapter 7 Principles of bacterial pathogenesis". in Bittar, Neville, E, B. Microbiology. Elsevier. p. 87. ISBN 978-1-55938-814-6.
- ↑ Rhinehart E; Friedman M (1999). Infection control in home care. Jones & Bartlett Learning. p. 11. ISBN 978-0-8342-1143-8. https://archive.org/details/infectioncontrol0000rhin/page/11.
- ↑ Roberts RJ, "Fish pathology, 3rd Edition", Elsevier Health Sciences, 2001.
- ↑ Roizman, B (1996). "42 Multiplication". in Baron, S. Medical Microbiology (4th ed.). University of Texas Medical Branch at Galveston. ISBN 978-0-9631172-1-2. https://www.ncbi.nlm.nih.gov/books/NBK8181/.
- ↑ Hilleman M (October 2004). "Strategies and mechanisms for host and pathogen survival in acute and persistent viral infections". Proceedings of the National Academy of Sciences of the United States of America 101 (Suppl 2): 14560–14566. doi:10.1073/pnas.0404758101. PMID 15297608. Bibcode: 2004PNAS..10114560H.
- ↑ Greggs WM; Clouser CL; Patterson SE; Manksy LM (April 2012). "Discovery of drugs that possess activity against feline leukemia virus". Journal of General Virology 93 (4): 900–905. doi:10.1099/vir.0.039909-0. PMID 22258856. PMC 3542715. http://jgv.sgmjournals.org/content/93/Pt_4/900.full.
- ↑ 34.0 34.1 Nester E; Anderson D; Evans Roberts, C; Nester M (2009). Microbiology: A human perspective. McGraw Hill. pp. 336–337. ISBN 978-1-55938-814-6.
- ↑ Møller M; El Maghrabi R; Olesen N; Thomsen VØ (November 2004). "Safe inoculation of blood and bone marrow for liquid culture detection of mycobacteria". Occupational Medicine 54 (8): 530–3. doi:10.1093/occmed/kqh106. PMID 15520021.
- ↑ Madigan MT (2009) Brock Biology of Microorganisms: Pearson/Benjamin Cummings.
- ↑ (Boseck 1982) (Slonczewski & Foster 2017)
- ↑ 38.0 38.1 Mackay I (2007). Real-time PCR in Microbiology: From Diagnosis to Characterisation. Horizon Scientific Press. pp. 1–25. ISBN 9781904455189.
- ↑ 39.0 39.1 Viljoen GJ, ed (2005). Molecular Diagnostic PCR Handbook. Springer. p. 58. ISBN 978-1-4020-3404-6.
- ↑ Tang YW; Persing DH (2009). Encyclopedia of Microbiology. Oxford Academic Press. pp. 308–320. ISBN 978-0-12-373944-5.
- ↑ WHO (April 2014). "Antimicrobial resistance: global report on surveillance 2014". WHO. https://www.who.int/drugresistance/documents/surveillancereport/en/.
- ↑ Vickery K, Hu H, Jacombs AS, Bradshaw DA, Deva AK (2013) A review of bacterial biofilms and their role in device-associated infection. Healthcare Infection .
- ↑ Stewart PS; Costerton JW (July 2001). "Antibiotic resistance of bacteria in biofilms". Lancet 358 (9276): 135–8. doi:10.1016/S0140-6736(01)05321-1. PMID 11463434. https://scholarworks.montana.edu/xmlui/handle/1/13570.
- ↑ (Gordillo Altamirano & Barr 2019) (Kortright et al. 2019)
- ↑ Taguchi T, Yabe M, Odaki H, Shinozaki M, Metsä-Ketelä M, Arai T, Okamoto S, Ichinose K (2013) Biosynthetic Conclusions from the Functional Dissection of Oxygenases for Biosynthesis of Actinorhodin and Related Streptomyces Antibiotics. Chemistry & Biology 20:510-520.
- ↑ Williams NT (2010) Probiotics. American Journal of Health-System Pharmacy 67:449-458.
External links
 |

