Biology:Transaldolase
| Transaldolase | |||||||||
|---|---|---|---|---|---|---|---|---|---|
| Identifiers | |||||||||
| EC number | 2.2.1.2 | ||||||||
| CAS number | 9014-46-4 | ||||||||
| Databases | |||||||||
| IntEnz | IntEnz view | ||||||||
| BRENDA | BRENDA entry | ||||||||
| ExPASy | NiceZyme view | ||||||||
| KEGG | KEGG entry | ||||||||
| MetaCyc | metabolic pathway | ||||||||
| PRIAM | profile | ||||||||
| PDB structures | RCSB PDB PDBe PDBsum | ||||||||
| Gene Ontology | AmiGO / QuickGO | ||||||||
| |||||||||
| Transaldolase | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
 | |||||||||||
| Identifiers | |||||||||||
| Symbol | Transaldolase | ||||||||||
| Pfam | PF00923 | ||||||||||
| InterPro | IPR001585 | ||||||||||
| PROSITE | PDOC00741 | ||||||||||
| SCOP2 | 1ucw / SCOPe / SUPFAM | ||||||||||
| |||||||||||
| transaldolase 1 | |
|---|---|
| Identifiers | |
| Symbol | TALDO1 |
| NCBI gene | 6888 |
| HGNC | 11559 |
| OMIM | 602063 |
| RefSeq | NM_006755 |
| UniProt | P37837 |
| Other data | |
| EC number | 2.2.1.2 |
| Locus | Chr. 11 p15.5-15.4 |
| transaldolase B | |
|---|---|
| Identifiers | |
| Symbol | talB |
| NCBI gene | 4199095 |
| PDB | 1onr |
| RefSeq | NC_008245.1 |
| UniProt | P0A870 |
| Other data | |
| EC number | 2.2.1.2 |
Transaldolase is an enzyme (EC 2.2.1.2) of the non-oxidative phase of the pentose phosphate pathway. In humans, transaldolase is encoded by the TALDO1 gene.[3][4]
The following chemical reaction is catalyzed by transaldolase:
Clinical significance
The pentose phosphate pathway has two metabolic functions: (1) generation of nicotinamide adenine dinucleotide phosphate (reduced NADPH), for reductive biosynthesis, and (2) formation of ribose, which is an essential component of ATP, DNA, and RNA. Transaldolase links the pentose phosphate pathway to glycolysis. In patients with deficiency of transaldolase, there's an accumulation of erythritol (from erythrose 4-phosphate), D-arabitol, and ribitol.[5][6]
The deletion in 3 base pairs in the TALDO1 gene results in the absence of serine at position 171 of the transaldolase protein, which is part of a highly conserved region, suggesting that the mutation causes the transaldolase deficiency that is found in erythrocytes and lymphoblasts.[5] The deletion of this amino acid can lead to liver cirrhosis and hepatosplenomegaly (enlarged spleen and liver) during early infancy. Transaldolase is also a target of autoimmunity in patients with multiple sclerosis.[7]
Structure
Transaldolase is a single domain composed of 337 amino acids. The core structure is an α/β barrel, similar to other class I aldolases, made up of eight parallel β-sheets and seven α-helices. There are also seven additional α-helices that are not part of the barrel. Hydrophobic amino acids are located between the β-sheets in the barrel and the surrounding α-helices to contribute to packing, such as the area containing Leu-168, Phe-170, Phe-189, Gly-311, and Phe-315. In the crystal, human transaldolase forms a dimer, with the two subunits connected by 18 residues in each subunit. See mechanism to the left for details.
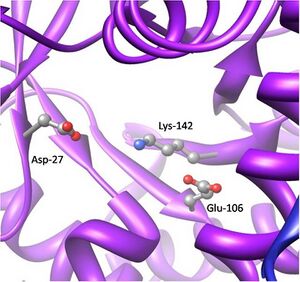
The active site, located in the center of the barrel, contains three key residues: lysine-142, glutamate-106, and aspartate-27. The lysine holds the sugar in place while the glutamate and aspartate act as proton donors and acceptors.[1]
Mechanism of catalysis
The residue of lysine-142 in the active site of transaldolase forms a Schiff base with the keto group in sedoheptulose-7-phosphate after deprotonation by another active site residue, glutamate-106. The reaction mechanism is similar to the reverse reaction catalyzed by aldolase: The bond joining carbons 3 and 4 is broken, leaving dihydroxyacetone joined to the enzyme via a Schiff base. This cleavage reaction generates the unusual aldose sugar erythrose-4-phosphate. Then transaldolase catalyzes the condensation of glyceraldehyde-3-phosphate with the Schiff base of dihydroxyacetone, yielding enzyme-bound fructose 6-phosphate. Hydrolysis of the Schiff base liberates free fructose 6-phosphate, one of the products of the pentose phosphate pathway.
See also
- Transaldolase deficiency
References
- ↑ 1.0 1.1 1.2 PDB: 1F05; "The three-dimensional structure of human transaldolase". FEBS Lett. 475 (3): 205–8. June 2000. doi:10.1016/S0014-5793(00)01658-6. PMID 10869557.
- ↑ Molecular graphics images were produced using the UCSF Chimera package from the Resource for Biocomputing, Visualization, and Informatics at the University of California, San Francisco. "UCSF Chimera–a visualization system for exploratory research and analysis". J Comput Chem 25 (13): 1605–12. October 2004. doi:10.1002/jcc.20084. PMID 15264254.
- ↑ "Entrez Gene: transaldolase 1". https://www.ncbi.nlm.nih.gov/sites/entrez?Db=gene&Cmd=ShowDetailView&TermToSearch=6888.
- ↑ "The human transaldolase gene (TALDO1) is located on chromosome 11 at p15.4-p15.5". Genomics 45 (1): 233–8. October 1997. doi:10.1006/geno.1997.4932. PMID 9339383.
- ↑ 5.0 5.1 5.2 "Transaldolase deficiency: liver cirrhosis associated with a new inborn error in the pentose phosphate pathway". Am. J. Hum. Genet. 68 (5): 1086–92. May 2001. doi:10.1086/320108. PMID 11283793.
- ↑ Perl A (June 2007). "The pathogenesis of transaldolase deficiency". IUBMB Life 59 (6): 365–73. doi:10.1080/15216540701387188. PMID 17613166.
- ↑ "Evaluation of autoimmunity to transaldolase in multiple sclerosis". Autoimmunity. 102. 2004. 155–71. doi:10.1385/1-59259-805-6:155. ISBN 978-1-59259-805-2.
- ↑ "Crystal structure of the reduced Schiff-base intermediate complex of transaldolase B from Escherichia coli: mechanistic implications for class I aldolases". Protein Sci. 6 (1): 119–24. January 1997. doi:10.1002/pro.5560060113. PMID 9007983. PMC 2143518. http://www.proteinscience.org/cgi/content/abstract/6/1/119.
External links
- Transaldolase at the US National Library of Medicine Medical Subject Headings (MeSH)
 |
![Reaction scheme for the conversion of sedoheptulose-7-phosphate to fructose-6-phosphate.[8]](/wiki/images/thumb/d/d3/Transaldolasemech.jpg/267px-Transaldolasemech.jpg)
![The pentose phosphate pathway adapted from (Verhoeven, 2001)[5]](/wiki/images/thumb/2/29/Transaldolaserepofppp.jpg/250px-Transaldolaserepofppp.jpg)
