Biology:Tudor domain
| TUDOR domain | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
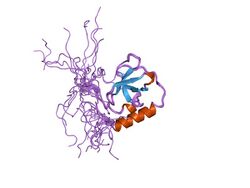 Structure of a TUDOR domain. | |||||||||||
| Identifiers | |||||||||||
| Symbol | TUDOR | ||||||||||
| Pfam | PF00567 | ||||||||||
| Pfam clan | CL0049 | ||||||||||
| InterPro | IPR008191 | ||||||||||
| SMART | TUDOR | ||||||||||
| PROSITE | PDOC50304 | ||||||||||
| SCOP2 | 3fdr / SCOPe / SUPFAM | ||||||||||
| CDD | cd04508 | ||||||||||
| |||||||||||
In molecular biology, a Tudor domain is a conserved protein structural domain originally identified in the Tudor protein encoded in Drosophila.[1] The Tudor gene was found in a Drosophila screen for maternal factors that regulate embryonic development or fertility.[2] Mutations here are lethal for offspring, inspiring the name Tudor, as a reference to the Tudor King Henry VIII and the several miscarriages experienced by his wives.[1]
Structure
A Tudor domain is a protein region approximately 60 amino acids in length, which folds into an SH3-like structure with a five-stranded antiparallel beta-barrel form.[1] Tudor domains can further be organized into functional units consisting of either a single Tudor domain, tandem Tudor domains, or hybrid Tudor domains consisting of two Tudor domains linked by an anti-parallel beta-sheet made from their shared second and third beta-strands.[1] An essential component of the Tudor domain structure is the aromatic-binding cage formed by several (typically 4–5) aromatic amino acid residues.[1]
Interaction with methylated residues
Tudor domains exert their functions by recognizing and binding methylated lysine and arginine residues, allowing them to function as histone readers in an epigenetic context.[1] This occurs through cation–pi interactions between the methylated Arg/Lys residue and the aromatic residues of the Tudor domain's aromatic-binding cage.[1] Depending on the Tudor domain, this interaction can be methylation state-specific (mono-, di-, or trimethylation).[1]
Function
DNA transcription and modification
Tudor domain proteins are involved in epigenetic regulation and can alter transcription by recognizing post-translational histone modifications and as adaptor proteins.[2] Recognition of methylated arginine and lysine histone residues results in the recruitment of downstream effectors, leading to chromatin silencing or activation depending on the Tudor domain protein and context.[1] For example, the human TDRD3 protein binds methylated arginine residues and promotes transcription of estrogen-responsive elements.[3] Conversely, the Polycomb-like protein (PCL) acts as an adaptor to recruit components of the Polycomb repressive complex 2 (PRC2), a histone H3K27 methyltransferase that represses transcription.[4] Additionally, Tudor domain proteins can repress transcription by recruiting DNA-methyltransferases to promote DNA methylation and heterochromatin assembly.[1] Tudor domain proteins also have the function of maintaining and propagating epigenetic modifications.[1]
Genome stability
The Tudor domain is involved in the silencing of selfish genetic elements, such as retrotransposons.[5] This functionality is performed both directly through Tudor-containing proteins, such as Tdrd7, as well as through piRNA synthesis.[6] Tudor domains are essential in the localization of protein machinery involved in piRNA creation, such as localization of Yb protein to the Yb body, assembly of the pole plasm in Drosophila, and recruitment of proteins to load Piwi with piRNA.[5]
DNA damage response
The human p53-binding protein 1 (TRP53BP1) is a Tudor domain protein involved in the DNA damage response (DDR) pathway, which functions to protect the genome from external stimuli.[5] It is a cascade of events that senses damage through adaptor proteins and triggers responses including cell cycle arrest, DNA repair, transcriptional modifications, and apoptosis.[5] TRP53BP1s Tudor domain mediates binding to sensors that accumulate at the sites of damage, and also functions as the adaptor promoting effector recruitment to the damaged sites.[5] TRP53BP1 is essential for DDR as it plays a very complex role in the regulation and recruitment of multiple other proteins involved.[5]
RNA metabolism
Tudor domain proteins involved in RNA metabolism have an extended Tudor domain of approximately 180 amino acids.[5] These proteins contain RNA-binding motifs to target RNAs, or bind to dimethylated arginines of proteins bound to RNAs.[5] These proteins regulate multiple aspects of RNA metabolism, including processing, stability, translation, and small RNA pathways.[5] Specifically, the survival motor neuron (SMN) protein is a Tudor domain protein that mediates the assembly of snRNPs (small nuclear ribonucleoproteins), by binding snRNAs and recruiting asymmetrically dimethylated arginines of SM proteins that form the protein constituent of snRNPs.[5] SMN promotes the maturation of snRNPs, which are essential for spliceosome assembly and intron removal.[5]
Examples
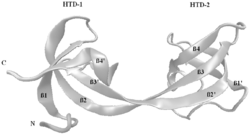
The proteins TP53BP1 (Tumor suppressor p53-binding protein 1) and its fission yeast homolog Crb2[8] and JMJD2A (Jumonji domain containing 2A) contain either tandem or double Tudor domains and recognize methylated histones.[9][10]
The structurally characterized Tudor domain in human SMN (survival of motor neuron) is a strongly bent anti-parallel β-sheet consisting of five β-strands with a barrel-like fold and recognizes symmetrically dimethylated arginine.[11]
Other Tudor domain containing proteins include AKAP1 (A-kinase anchor protein 1)[12] and ARID4A (AT rich interactive domain 4A) among others. A well known Tudor domain containing protein is Staphylococcal Nuclease Domain Containing 1 (SND1)/Tudor-SN/p100 co activator.[13] SND1 is involved in RISC complex and interacts with AEG-1 oncogene.[14] SND1 is also acts as an oncogene and plays a very important role in HCC and colon cancer.[15] The SND1 Tudor domain binds to methylated arginine in the PIWIL1 protein.[16] Tudor containing SND1 promotes tumor angiogenesis in human hepatocellular carcinoma through a novel pathway which involves NF-kappaB and miR-221.[17] Tudor SND1 is also present in the Drosophila melanogaster.[6]
References
- ↑ 1.00 1.01 1.02 1.03 1.04 1.05 1.06 1.07 1.08 1.09 1.10 "Tudor Domains as Methyl-Lysine and Methyl-Arginine Readers". Chromatin Signaling and Diseases. Elsevier. 2016. pp. 149–165. doi:10.1016/b978-0-12-802389-1.00008-3. ISBN 978-0-12-802389-1.
- ↑ 2.0 2.1 "Tudor: a versatile family of histone methylation 'readers'". Trends in Biochemical Sciences 38 (11): 546–55. November 2013. doi:10.1016/j.tibs.2013.08.002. PMID 24035451.
- ↑ "TDRD3 is an effector molecule for arginine-methylated histone marks". Molecular Cell 40 (6): 1016–23. December 2010. doi:10.1016/j.molcel.2010.11.024. PMID 21172665.
- ↑ "An H3K36 methylation-engaging Tudor motif of polycomb-like proteins mediates PRC2 complex targeting". Molecular Cell 49 (3): 571–82. February 2013. doi:10.1016/j.molcel.2012.11.026. PMID 23273982.
- ↑ 5.00 5.01 5.02 5.03 5.04 5.05 5.06 5.07 5.08 5.09 5.10 "Tudor domain proteins in development". Development 139 (13): 2255–66. July 2012. doi:10.1242/dev.073304. PMID 22669818.
- ↑ 6.0 6.1 "Tudor domain-containing proteins of Drosophila melanogaster". Development, Growth & Differentiation 54 (1): 32–43. January 2012. doi:10.1111/j.1440-169x.2011.01308.x. PMID 23741747.
- ↑ "Molecular recognition of H3/H4 histone tails by the tudor domains of JMJD2A: a comparative molecular dynamics simulations study". PLOS ONE 6 (3): e14765. March 2011. doi:10.1371/journal.pone.0014765. PMID 21464980. Bibcode: 2011PLoSO...614765O.
- ↑ "Structural basis for the methylation state-specific recognition of histone H4-K20 by 53BP1 and Crb2 in DNA repair". Cell 127 (7): 1361–73. December 2006. doi:10.1016/j.cell.2006.10.043. PMID 17190600.
- ↑ "Recognition of histone H3 lysine-4 methylation by the double tudor domain of JMJD2A". Science 312 (5774): 748–51. May 2006. doi:10.1126/science.1125162. PMID 16601153. Bibcode: 2006Sci...312..748H.
- ↑ "Distinct binding modes specify the recognition of methylated histones H3K4 and H4K20 by JMJD2A-tudor". Nature Structural & Molecular Biology 15 (1): 109–11. January 2008. doi:10.1038/nsmb1326. PMID 18084306.
- ↑ "High-resolution X-ray and NMR structures of the SMN Tudor domain: conformational variation in the binding site for symmetrically dimethylated arginine residues". Journal of Molecular Biology 327 (2): 507–20. March 2003. doi:10.1016/s0022-2836(03)00148-7. PMID 12628254.
- ↑ "The KH-Tudor domain of a-kinase anchoring protein 149 mediates RNA-dependent self-association". Biochemistry 45 (50): 14980–9. December 2006. doi:10.1021/bi061418y. PMID 17154535.
- ↑ "A micrococcal nuclease homologue in RNAi effector complexes". Nature 425 (6956): 411–4. September 2003. doi:10.1038/nature01956. PMID 14508492. Bibcode: 2003Natur.425..411C.
- ↑ "Increased RNA-induced silencing complex (RISC) activity contributes to hepatocellular carcinoma". Hepatology 53 (5): 1538–48. May 2011. doi:10.1002/hep.24216. PMID 21520169.
- ↑ "Astrocyte elevated gene-1 (AEG-1): A multifunctional regulator of normal and abnormal physiology". Pharmacology & Therapeutics 130 (1): 1–8. April 2011. doi:10.1016/j.pharmthera.2011.01.008. PMID 21256156.
- ↑ "Structural basis for recognition of arginine methylated Piwi proteins by the extended Tudor domain". Proceedings of the National Academy of Sciences of the United States of America 107 (43): 18398–403. October 2010. doi:10.1073/pnas.1013106107. PMID 20937909. Bibcode: 2010PNAS..10718398L.
- ↑ "Multifunction protein staphylococcal nuclease domain containing 1 (SND1) promotes tumor angiogenesis in human hepatocellular carcinoma through novel pathway that involves nuclear factor κB and miR-221". The Journal of Biological Chemistry 287 (17): 13952–8. April 2012. doi:10.1074/jbc.M111.321646. PMID 22396537.
 |

