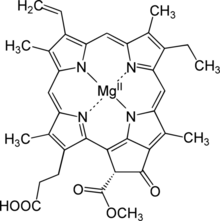
Chemistry:Protochlorophyllide
| |
| Names | |
|---|---|
| IUPAC name
Magnesium (21R)-3-(2-carboxyethyl)-14-ethyl-21-(methoxycarbonyl)-4,8,13,18-tetramethyl-20-oxo-9-vinyl-3,4,23,25-tetradehydrophorbine-23,25-diide
| |
| Other names
Monovinyl protochlorophyllide
| |
| Identifiers | |
3D model (JSmol)
|
|
| ChEBI | |
| ChemSpider | |
| KEGG | |
PubChem CID
|
|
| |
| |
| Properties | |
| C35H32MgN4O5 | |
| Molar mass | 612.957 g/mol |
Except where otherwise noted, data are given for materials in their standard state (at 25 °C [77 °F], 100 kPa). | |
| Infobox references | |
Protochlorophyllide,[1] or monovinyl protochlorophyllide, is an intermediate in the biosynthesis of chlorophyll a. It lacks the phytol side-chain of chlorophyll and the reduced pyrrole in ring D.[2] Protochlorophyllide is highly fluorescent; mutants that accumulate it glow red if irradiated with blue light.[3] In angiosperms, the later steps which convert protochlorophyllide to chlorophyll are light-dependent, and such plants are pale (chlorotic) if grown in the darkness. Gymnosperms, algae, and photosynthetic bacteria have another, light-independent enzyme and grow green in the darkness as well.
Conversion to chlorophyll
The enzyme that converts protochlorophyllide to chlorophyllide a, the next intermediate on the biosynthetic pathway,[4] is protochlorophyllide reductase,[5] EC 1.3.1.33. There are two structurally unrelated proteins with this activity: the light-dependent and the dark-operative. The light-dependent reductase needs light to operate. The dark-operative version is a completely different protein, consisting of three subunits that exhibit significant sequence similarity to the three subunits of nitrogenase, which catalyzes the formation of ammonia from dinitrogen.[6] This enzyme might be evolutionary older but (being similar to nitrogenase) is highly sensitive to free oxygen and does not work if its concentration exceeds about 3%.[7] Hence, the alternative, light-dependent version needed to evolve.
Most of the photosynthetic bacteria have both light-dependent and light-independent reductases. Angiosperms have lost the dark-operative form and rely on 3 slightly different copies of light-dependent version, frequently abbreviated as POR A, B, and C. Gymnosperms have much more copies of the similar gene (Loblolly pine has about 11 Loblolly Pine (Pinus taeda L.) Contains Multiple Expressed Genes Encoding Light-Dependent NADPH:Protochlorophyllide Oxidoreductase (POR)). In plants, POR is encoded in the cell nucleus and only later transported to its place of work, chloroplast. Unlike with POR, in plants and algae that have the dark-operative enzyme it is at least partially encoded in the chloroplast genome.[8]
Potential danger for plant
Chlorophyll itself is bound to proteins and can transfer the absorbed energy in the required direction. Protochlorophyllide, however, occurs mostly in the free form and, under light conditions, acts as a photosensitizer, forming highly toxic free radicals. Hence, plants need an efficient mechanism of regulating the amount of chlorophyll precursor. In angiosperms, this is done at the step of δ-aminolevulinic acid (ALA), one of the intermediate compounds in the biosynthetic pathway. Plants that are fed by ALA accumulate high and toxic levels of protochlorophyllide, as do mutants with a disrupted regulatory system.
Arabidopsis FLU mutant with damaged regulation can survive only either in a continuous darkness (protochlorophyllide is not dangerous in the darkness) or under continuous light, when the plant is can convert all produced protochlorophyllide into chlorophyll and does not over accumulate it despite the lack of regulation. In barley Tigrina mutant (mutated on the same gene,[9]) light kills the majority of the leaf tissue that has developed in the darkness, but part of the leaf that originated during the day survives. As a result, the leaves are covered by white stripes of necrotic regions, and the number of the white stripes is close to the age of the leaf in days. Green regions survive the subsequent nights, likely because the synthesis of chlorophyll in the mature leaf tissue is greatly reduced anyway.
Biosynthesis regulatory protein FLU
In spite of numerous past attempts to find the mutant that overacumulates protochlorophyllide under usual conditions, only one such gene (flu) is currently (2009) known. Flu (first described in [3]) is a nuclear-encoded, chloroplast-located protein that appears containing only protein-protein interaction sites. It is currently not known which other proteins interact through this linker. The regulatory protein is a transmembrane protein that is located in the thylakoid membrane. Later, it was discovered that Tigrina mutants in barley, known a long time ago, are also mutated in the same gene.[9] It is not obvious why no mutants of any other gene were observed; maybe mutations in other proteins, involved into the regulatory chain, are fatal. Flu is a single gene, not a member of the gene family.
Later, by the sequence similarity, a similar protein was found in Chlamydomonas algae,[10] showing that this regulatory subsystem existed a long time before the angiosperms lost the independent conversion enzyme. In a different manner, the Chlamydomonas regulatory protein is more complex: It is larger, crosses the thylakoid membrane twice rather than once, contains more protein-protein interactions sites, and even undergoes alternative splicing. It appears that the regulatory system underwent simplification during evolution.
References
- ↑ KEGG compound database entry [1]
- ↑ Willows, Robert D. (2003). "Biosynthesis of chlorophylls from protoporphyrin IX". Natural Product Reports 20 (6): 327–341. doi:10.1039/B110549N. PMID 12828371.
- ↑ 3.0 3.1 Meskauskiene R, Nater M, Goslings D, Kessler F, op den Camp R, Apel K. FLU: a negative regulator of chlorophyll biosynthesis in Arabidopsis thaliana. Proceedings of the National Academy of Sciences of the United States of America. 2001; 98(22):12826-31 pdf.
- ↑ R. Caspi (2007-07-18). "3,8-divinyl-chlorophyllide a biosynthesis I (aerobic, light-dependent)". MetaCyc Metabolic Pathway Database. https://biocyc.org/META/NEW-IMAGE?type=PATHWAY&object=CHLOROPHYLL-SYN. Retrieved 2020-06-04.
- ↑ KEGG enzyme entry 1.3.1.33 [2]
- ↑ Yuichi FujitaDagger and Carl E. Bauer (2000). Reconstitution of Light-independent Protochlorophyllide Reductase from Purified Bchl and BchN-BchB Subunits. J. Biol. Chem., Vol. 275, Issue 31, 23583-23588. [3]
- ↑ S.Yamazaki, J.Nomata, Y.Fujita (2006) Differential operation of dual protochlorophyllide reductases for chlorophyll biosynthesis in response to environmental oxygen levels in the cyanobacterium Leptolyngbya boryana. Plant Physiology, 2006, 142, 911-922 [4]
- ↑ J Li, M Goldschmidt-Clermont, M P Timko (1997). Chloroplast-encoded chlB is required for light-independent protochlorophyllide reductase activity in Chlamydomonas reinhardtii. Plant Cell 5(12): 1817–1829. [5].
- ↑ 9.0 9.1 Lee, Keun Pyo; Kim, Chanhong; Lee, Dae Won; Apel, Klaus (2003). "TIGRINA d, required for regulating the biosynthesis of tetrapyrroles in barley, is an ortholog of the FLU gene of Arabidopsis thaliana". FEBS Letters 553 (1–2): 119–124. doi:10.1016/s0014-5793(03)00983-9. PMID 14550558.
- ↑ A Falciatore, L Merendino, F Barneche, M Ceol, R Meskauskiene, K Apel, JD Rochaix (2005). The FLP proteins act as regulators of chlorophyll synthesis in response to light and plastid signals in Chlamydomonas. Genes & Dev, 19:176-187 [6]
