Chemistry:Cofactor F430
| Error creating thumbnail: Unable to save thumbnail to destination | |
| Identifiers | |
|---|---|
3D model (JSmol)
|
|
| ChEBI | |
| ChemSpider | |
PubChem CID
|
|
| |
| |
| Properties | |
| C42H51N6NiO−13 | |
| Molar mass | 906.58014 |
| Appearance | Yellow solid |
Except where otherwise noted, data are given for materials in their standard state (at 25 °C [77 °F], 100 kPa). | |
| Infobox references | |
F430 is the cofactor (sometimes called the coenzyme) of the enzyme methyl coenzyme M reductase (MCR).[1][2] MCR catalyzes the reaction EC 2.8.4.1 that releases methane in the final step of methanogenesis:
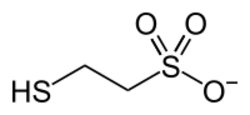 |
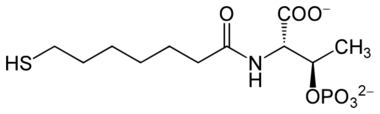 |
It is found only in methanogenic Archaea[3] and anaerobic methanotrophic Archaea. It occurs in relatively high concentrations in archaea that are involved in reverse methanogenesis: these can contain up to 7% by weight of the nickel protein.[4]
Structure
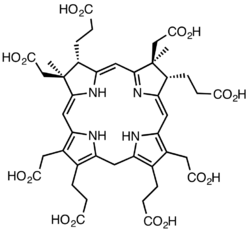
The trivial name cofactor F430 was assigned in 1978 based on the properties of a yellow sample extracted from Methanobacterium thermoautotrophicum, which had a spectroscopic maximum at 430 nm.[5] It was identified as the MCR cofactor in 1982[6] and the complete structure was deduced by X-ray crystallography and NMR spectroscopy.[7] Coenzyme F430 features a reduced porphyrin in a macrocyclic ring system called a corphin.[8] In addition, it possesses two additional rings in comparison to the standard tetrapyrrole (rings A-D), having a γ-lactam ring E and a keto-containing carbocyclic ring F. It is the only natural tetrapyrrole containing nickel, an element rarely found in biological systems.[9]
Biosynthesis
 |
 |
 |
The biosynthesis builds from uroporphyrinogen III, the progenitor of all natural tetrapyrroles, including chlorophyll, vitamin B12, phycobilins, siroheme, heme, and heme d1. It is converted to sirohydrochlorin via dihydrosirohydrochlorin.[10] Insertion of nickel into this tetrapyrrole is catalysed in reaction EC 4.99.1.11 by the same chelatase, CbiX, which inserts cobalt in the biosynthesis of cobalamin, here giving nickel(II)-sirohydrochlorin.[11]

The ATP-dependent Ni-sirohydrochlorin a,c-diamide synthase (CfbE) then converts the a and c acetate side chains to acetamide in reactions EC 6.3.5.12, generating nickel(II)-sirohydrochlorin a,c-diamide. The sequence of the two amidations is random.[11] A two-component complex Ni-sirohydrochlorin a,c-diamide reductive cyclase (CfbCD) carries out a 6-electron and 7-proton reduction of the ring system in a reaction EC 6.3.3.7 generating the 15,173-seco-F430-173-acid (seco-F430) intermediate. Reduction involves ATP hydrolysis and electrons are relayed through two 4Fe-4S centres. In the final step, the keto-containing carbocyclic ring F is formed by an ATP-dependent enzyme Coenzyme F(430) synthetase (CfbB) in reaction EC 6.4.1.9, generating coenzyme F430.[11][12][13] This enzyme is a MurF-like ligase, as found in peptidoglycan biosynthesis.
References
- ↑ Stephen W., Ragdale (2014). "Biochemistry of Methyl-Coenzyme M Reductase: The Nickel Metalloenzyme that Catalyzes the Final Step in Synthesis and the First Step in Anaerobic Oxidation of the Greenhouse Gas Methane". in Peter M.H. Kroneck and Martha E. Sosa Torres. The Metal-Driven Biogeochemistry of Gaseous Compounds in the Environment. Metal Ions in Life Sciences. 14. Springer. pp. 125–145. doi:10.1007/978-94-017-9269-1_6. ISBN 978-94-017-9268-4.
- ↑ Hofer, Ursula (2016). "Masters of methane". Nature Reviews Microbiology 14 (12): 727. doi:10.1038/nrmicro.2016.170. PMID 27818502.
- ↑ Thauer RK (1998). "Biochemistry of Methanogenesis: a Tribute to Marjory Stephenson". Microbiology 144 (9): 2377–2406. doi:10.1099/00221287-144-9-2377. PMID 9782487.
- ↑ "A conspicuous nickel protein in microbial mats that oxidize methane anaerobically". Nature 426 (6968): 878–81. December 2003. doi:10.1038/nature02207. PMID 14685246. Bibcode: 2003Natur.426..878K.
- ↑ Gunsalus, R.P.; Wolfe, R.S. (1978). "Chromophoric factors F342 and F430 of Methanobacterium thermoautotrophicum". FEMS Microbiology Letters 3 (4): 191–193. doi:10.1111/j.1574-6968.1978.tb01916.x.
- ↑ Ellefson, W. L.; Whitman, W. B.; Wolfe, R. S. (1982). "Nickel-containing factor F430: Chromophore of the methylreductase of Methanobacterium". Proceedings of the National Academy of Sciences 79 (12): 3707–3710. doi:10.1073/pnas.79.12.3707. PMID 6954513. Bibcode: 1982PNAS...79.3707E.
- ↑ "Coenzyme F430 from Methanogenic Bacteria : Complete Assignment of Configuration Based on an X-ray Analysis of 12,13-diepi-F430 Pentamethyl Ester and on NMR Spectroscopy". Helvetica Chimica Acta 74 (4): 697–716. 1991. doi:10.1002/hlca.19910740404.
- ↑ Eschenmoser, A. (1986). "Chemistry of Corphinoids". Annals of the New York Academy of Sciences 471 (1 International): 108–129. doi:10.1111/j.1749-6632.1986.tb48030.x. Bibcode: 1986NYASA.471..108E.
- ↑ Johnson, Michael K.; Scott, Robert A. (2 October 2017). Metalloprotein Active Site Assembly. Wiley. ISBN 9781119159834.
- ↑ Mucha, Helmut; Keller, Eberhard; Weber, Hans; Lingens, Franz; Trösch, Walter (1985-10-07). "Sirohydrochlorin, a precursor of factor F430 biosynthesis in Methanobacterium thermoautotrophicum". FEBS Letters 190 (1): 169–171. doi:10.1016/0014-5793(85)80451-8.
- ↑ 11.0 11.1 11.2 Moore, Simon J.; Sowa, Sven T.; Schuchardt, Christopher; Deery, Evelyne; Lawrence, Andrew D.; Ramos, José Vazquez; Billig, Susan; Birkemeyer, Claudia et al. (2017). "Elucidation of the biosynthesis of the methane catalyst coenzyme F430". Nature 543 (7643): 78–82. doi:10.1038/nature21427. PMID 28225763. Bibcode: 2017Natur.543...78M.
- ↑ Zheng, Kaiyuan; Ngo, Phong D.; Owens, Victoria L.; Yang, Xue-Peng; Mansoorabadi, Steven O. (2016). "The biosynthetic pathway of coenzyme F430 in methanogenic and methanotrophic archaea". Science 354 (6310): 339–342. doi:10.1126/science.aag2947. PMID 27846569. Bibcode: 2016Sci...354..339Z.
- ↑ R. Caspi (2017-01-09). "Pathway: factor 430 biosynthesis". MetaCyc Metabolic Pathway Database. https://biocyc.org/META/NEW-IMAGE?type=PATHWAY&object=PWY-5196.
 |
