Biology:Methane monooxygenase
| Particulate methane monooxygenase | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
 | |||||||||||
| Identifiers | |||||||||||
| Symbol | pMMO | ||||||||||
| Pfam | PF02461 | ||||||||||
| InterPro | IPR003393 | ||||||||||
| OPM superfamily | 23 | ||||||||||
| OPM protein | 1yew | ||||||||||
| |||||||||||
Methane monooxygenase (MMO) is an enzyme capable of oxidizing the C-H bond in methane as well as other alkanes.[1] Methane monooxygenase belongs to the class of oxidoreductase enzymes (EC 1.14.13.25).
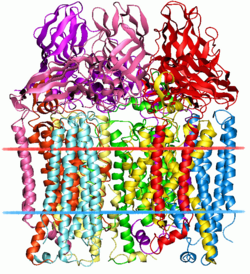
There are two forms of MMO: the well-studied soluble form (sMMO) and the particulate form (pMMO).[2] The active site in sMMO contains a di-iron center bridged by an oxygen atom (Fe-O-Fe), whereas the active site in pMMO utilizes copper. Structures of both proteins have been determined by X-ray crystallography; however, the location and mechanism of the active site in pMMO is still poorly understood and is an area of active research.
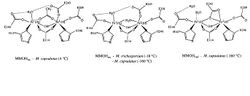
The particulate methane monooxygenase and related ammonia monooxygenase are integral membrane proteins, occurring in methanotrophs and ammonia oxidisers, respectively, which are thought to be related.[3] These enzymes have a relatively wide substrate specificity and can catalyse the oxidation of a range of substrates including ammonia, methane, halogenated hydrocarbons, and aromatic molecules.[4] These enzymes are homotrimers composed of 3 subunits - A (InterPro: IPR003393), B (InterPro: IPR006833) and C (InterPro: IPR006980) and most contain two monocopper centers.[5][6]
The A subunit from Methylococcus capsulatus (Bath) resides primarily within the membrane and consists of 7 transmembrane helices and a beta-hairpin, which interacts with the soluble region of the B subunit. A conserved glutamate residue is thought to contribute to a metal center.[5]
Methane monooxygenases are found in methanotrophic bacteria, a class of bacteria that exist at the interface of aerobic (oxygen-containing) and anaerobic (oxygen-devoid) environments. One of the more widely studied bacteria of this type is Methylococcus capsulatus (Bath). This bacterium was discovered in the hot springs of Bath, England.[7] Notably, strictly anaerobic methanotrophs may also harbour methane monooxygenases, although there are critical mismatches in the gene which prevent common methanotroph-seeking primers from matching.[8]
Soluble methane monooxygenase systems
Methanotrophic bacteria play an essential role of cycling carbon through anaerobic sediments. The chemistry behind the cycling takes a chemically inert hydrocarbon, methane, and converts it to a more active species, methanol. Other hydrocarbons are oxidized by MMOs, so a new hydroxylation catalyst based on the understanding of MMO systems could possibly make a more efficient use of the world supply of natural gas.[9]
This is a classic monooxygenase reaction in which two reducing equivalents from NAD(P)H are utilized to split the O-O bond of O2. One atom is reduced to water by a 2 e- reduction and the second is incorporated into the substrate to yield methanol:[10]
CH4 + O2 + NAD(P)H + H+ -> CH3OH + NAD(P)+ + H2O
Two forms of MMO have been found: soluble and particulate. The best characterized forms of soluble MMO contains three protein components: hydroxylase, the β unit, and the reductase. Each of which is necessary for effective substrate hydroxylation and NADH oxidation.[10]
Structure
X-ray crystallography of the MMO shows that it is a dimer formed of three subunits, α2β2γ2. With 2.2 A resolution, the crystallography shows that MMO is a relatively flat molecule with the dimensions of 60 x 100 x 120 A. In addition, there is a wide canyon running along the dimer interface with an opening in the center of the molecule. Most of the protomers involves helices from the α and β subunits with no participation from the γ subunit. Also, the interactions with the protomers resembles ribonucleotide reductase R2 protein dimer interaction, resembling a heart.[11][12] Each iron has a six coordinate octahedral environment. The dinuclear iron centers are positioned in the α subunit. Each iron atoms are also coordinated to a histidine δN atom, Fe 1 to a His 147 and Fe 2 to His 246, Fe 1 is a ligated to a monodentate carboxylate, Glu 114, a semi bridging carboxylate, Glu 144, and a water molecule.[9]
The substrate must bind near the active site in order for the reaction to take place. Near to the iron centers, there are hydrophobic pockets. It is thought that here the methane binds and is held until needed. From the X-ray crystallography, there is no direct path to these packets. However, a slight conformation change in the Phe 188 or The 213 side-chains could allow access.[9] This conformational change could be triggered by the binding of a coupling protein and the activase.
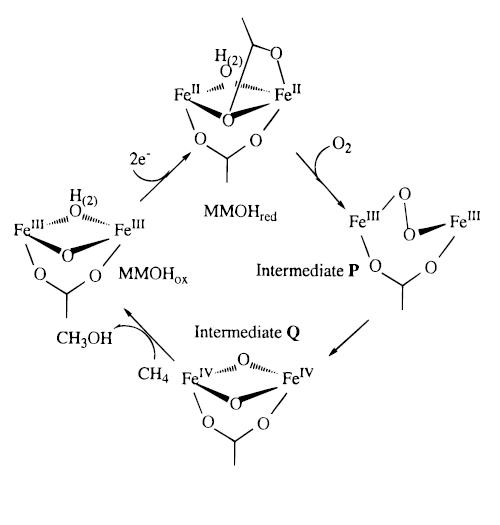
Upon reduction, one of the carboxylate ligands undergoes a “1,2 carboxylate” shift from behind a terminal monodentate ligand to a bridging ligand for the two irons, with the second oxygen coordinated to Fe 2. In the reduced form of MMOHred, the ligand environment for the Fe effectively becomes five coordinated, a form that permits the cluster to activate dioxygen.[10] The two irons are at this point oxidized to FeIV and have changed from low-spin ferromagnetic to high-spin antiferromagnetic.
Proposed catalytic cycle and mechanism
From the MMOHred, the diiron centers react with the O2 to form intermediate P. This intermediate is a peroxide species where the oxygens are bound symmetrically, suggested by spectroscopic studies.[13] However, the structure is not known. Intermediate P then converts to intermediate Q, which was proposed to contain two antiferromagnetically coupled high-spin FeIV centers.[10] This compound Q with its diamond core is critical to the oxidizing species for MMO.
There are two mechanisms suggested for the reaction between compound Q and the alkane: radical and nonradical. The radical mechanism starts with abstraction of the hydrogen atom from the substrate to form QH (the rate determining step), hydroxyl bridged compound Q and the free alkyl radical. The nonradical mechanism implies a concerted pathway, occurring via a four-center transition state and leading to a “hydrido-alkyl-Q” compound. As of 1999, the research suggests that the methane oxidation proceeds via a bound-radical mechanism.
It was suggested that the transition state for the radical mechanism involves a torsion motion of the hydroxyl OH ligand before the methyl radical can add to the bridging hydroxyl ligand to form the alcohol. As the radical approaches, the H atom of the alkane leave the coplanar tricoordinate O environment and bends upward to create a tetrahedral tetracoordinate O environment.[10]
The final step for this reaction is the elimination of the alcohol and the regeneration of the catalysts. There are a few ways in which this can occur. It could be a stepwise mechanism that starts with the elimination of the alcohol and an intermediate Fe-O-Fe core, and the latter can eliminate the water and regenerate the enzyme through a 2e- reduction. On the other hand, it can start with a 2e- reduction process of bridging the O1 atom to give a water molecule, followed by elimination of the alcohol and regeneration of the enzyme. In addition, it is possible that there is a concerted mechanism whereby the elimination of the methanol occurs spontaneously with 2e- reduction of the bridging O1 center and regeneration of the catalyst.[10]
See also
References
- ↑ "Chapter 6 Methane Monooxygenase: Functionalizing Methane at Iron and Copper". Sustaining Life on Planet Earth: Metalloenzymes Mastering Dioxygen and Other Chewy Gases. Metal Ions in Life Sciences. 15. Springer. 2015. pp. 205–256. doi:10.1007/978-3-319-12415-5_6. ISBN 978-3-319-12414-8.
- ↑ "A tale of two methane monooxygenases". Journal of Biological Inorganic Chemistry 22 (2–3): 307–319. April 2017. doi:10.1007/s00775-016-1419-y. PMID 27878395.
- ↑ "Evidence that particulate methane monooxygenase and ammonia monooxygenase may be evolutionarily related". FEMS Microbiology Letters 132 (3): 203–208. October 1995. doi:10.1111/j.1574-6968.1995.tb07834.x. PMID 7590173.
- ↑ "Molecular biology and biochemistry of ammonia oxidation by Nitrosomonas europaea". Archives of Microbiology 178 (4): 250–255. October 2002. doi:10.1007/s00203-002-0452-0. PMID 12209257. Bibcode: 2002ArMic.178..250A.
- ↑ 5.0 5.1 "Crystal structure of a membrane-bound metalloenzyme that catalyses the biological oxidation of methane". Nature 434 (7030): 177–182. March 2005. doi:10.1038/nature03311. PMID 15674245. Bibcode: 2005Natur.434..177L.
- ↑ "Particulate methane monooxygenase contains only mononuclear copper centers". Science 364 (6440): 566–570. May 2019. doi:10.1126/science.aav2572. PMID 31073062. Bibcode: 2019Sci...364..566R.
- ↑ "The acetylene reduction technique as an assay for nitrogenase activity in the methane oxidizing bacterium Methylococcus capsulatus strain bath". Archives of Microbiology 109 (1): 147–151. August 1976. doi:10.1007/BF00425127. Bibcode: 1976ArMic.109..147D.
- ↑ "pmoA Primers for detection of anaerobic methanotrophs" (in EN). Applied and Environmental Microbiology 77 (11): 3877–3880. June 2011. doi:10.1128/AEM.02960-10. PMID 21460105. Bibcode: 2011ApEnM..77.3877L.
- ↑ 9.0 9.1 9.2 "Crystal structure of a bacterial non-haem iron hydroxylase that catalyses the biological oxidation of methane". Nature 366 (6455): 537–543. December 1993. doi:10.1038/366537a0. PMID 8255292. Bibcode: 1993Natur.366..537R.
- ↑ 10.0 10.1 10.2 10.3 10.4 10.5 Basch, Harold (1999). "Mechanism of the Methane -> Methanol Conversion Reaction Catalyzed by Methane Monoxygenase: A Density Function Study". J. Am. Chem. Soc. 121 (31): 7249–7256. doi:10.1021/ja9906296.
- ↑ "Three-dimensional structure of the free radical protein of ribonucleotide reductase". Nature 345 (6276): 593–598. June 1990. doi:10.1038/345593a0. PMID 2190093. Bibcode: 1990Natur.345..593N.
- ↑ "Structure and function of the Escherichia coli ribonucleotide reductase protein R2". Journal of Molecular Biology 232 (1): 123–164. July 1993. doi:10.1006/jmbi.1993.1374. PMID 8331655.
- ↑ "Characterization of a Diiron(III) Peroxide Intermediate in the Reaction Cycle of Methane Monooxygenase Hydroxylase from Methylococcus capsulatus (Bath)". Journal of the American Chemical Society 117 (17): 4997–4998. 1995. doi:10.1021/ja00122a032.
Further reading
- The biological chemistry of the elements : the inorganic chemistry of life (2nd ed.). Oxford: Oxford University Press. 2008. ISBN 978-0-19-850848-9.
External links
- UMich Orientation of Proteins in Membranes protein/pdbid-1yew
- methane+monooxygenase at the US National Library of Medicine Medical Subject Headings (MeSH)
 |