Biology:Squalene monooxygenase
| Squalene epoxidase | |||||||||
|---|---|---|---|---|---|---|---|---|---|
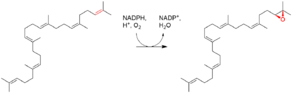 Chemical reaction catalyzed by squalene epoxidase. | |||||||||
| Identifiers | |||||||||
| EC number | 1.14.13.132 | ||||||||
| CAS number | 9029-62-3 | ||||||||
| Databases | |||||||||
| IntEnz | IntEnz view | ||||||||
| BRENDA | BRENDA entry | ||||||||
| ExPASy | NiceZyme view | ||||||||
| KEGG | KEGG entry | ||||||||
| MetaCyc | metabolic pathway | ||||||||
| PRIAM | profile | ||||||||
| PDB structures | RCSB PDB PDBe PDBsum | ||||||||
| Gene Ontology | AmiGO / QuickGO | ||||||||
| |||||||||
 Generic protein structure example |
Squalene monooxygenase (also called squalene epoxidase) is a eukaryotic enzyme that uses NADPH and diatomic oxygen to oxidize squalene to 2,3-oxidosqualene (squalene epoxide). Squalene epoxidase catalyzes the first oxygenation step in sterol biosynthesis and is thought to be one of the rate-limiting enzymes in this pathway.[1] In humans, squalene epoxidase is encoded by the SQLE gene.[2] Several eukaryote genomes lack a squalene monooxygenase encoding gene, but instead encode an alternative squalene epoxidase that performs the same task.[3]
Mechanism
The canonical squalene monooxygenase is a flavoprotein monooxygenase. Flavoprotein monooxygenase form flavin hydroperoxides at the enzyme active site, which then transfer the terminal oxygen atom of the hydroperoxide to the substrate. Squalene monooxygenase differs from other flavin monooxygenases in that the oxygen is inserted into the substrate as an epoxide rather than as a hydroxyl group. This enzyme contains a loosely bound FAD flavin and obtains electrons from NADPH-cytochrome P450 reductase, rather than binding NADPH directly. The alternative squalene epoxidase belongs to the fatty acid hydroxylase superfamily and obtains electrons from cytochrome b5.[3]
Inhibitors
Inhibitors of squalene epoxidase have found application mainly as antifungal drugs:[4]
Since squalene epoxidase is on the biosynthetic pathway leading to production of cholesterol, inhibitors of this enzyme may also find application in treatment of hypercholesterolemia.[6]
Localization
In baker's yeast (Saccharomyces cerevisiae), squalene epoxidase is localized to both the endoplasmic reticulum and lipid droplets. Only the ER localized protein is active.
Additional products
Squalene epoxidase also catalyzes the formation of diepoxysqualene (DOS). DOS is converted to 24(S),25-epoxylanosterol by lanosterol synthase.
See also
- Antifungal drug#Allylamines
References
- ↑ "Entrez Gene: SQLE squalene epoxidase". https://www.ncbi.nlm.nih.gov/sites/entrez?Db=gene&Cmd=ShowDetailView&TermToSearch=6713.
- ↑ "Localization of the squalene epoxidase gene (SQLE) to human chromosome region 8q24.1". Genomics 44 (1): 141–3. Aug 1997. doi:10.1006/geno.1997.4825. PMID 9286711.
- ↑ 3.0 3.1 "A widespread alternative squalene epoxidase participates in eukaryote steroid biosynthesis". Nature Microbiology 4 (2): 226–233. 2019. doi:10.1038/s41564-018-0305-5. PMID 30478288. https://biblio.ugent.be/publication/8587985.
- ↑ "Characterization of squalene epoxidase activity from the dermatophyte Trichophyton rubrum and its inhibition by terbinafine and other antimycotic agents". Antimicrobial Agents and Chemotherapy 40 (2): 443–7. Feb 1996. doi:10.1128/AAC.40.2.443. PMID 8834895.
- ↑ "Terbinafine: mode of action and properties of the squalene epoxidase inhibition". The British Journal of Dermatology 126 (Suppl 39): 2–7. Feb 1992. doi:10.1111/j.1365-2133.1992.tb00001.x. PMID 1543672.
- ↑ "Squalene epoxidase as hypocholesterolemic drug target revisited". Progress in Lipid Research 42 (1): 37–50. Jan 2003. doi:10.1016/S0163-7827(02)00029-2. PMID 12467639.
Further reading
- "Identifying leukocyte gene expression patterns associated with plasma lipid levels in human subjects". Atherosclerosis 191 (1): 63–72. Mar 2007. doi:10.1016/j.atherosclerosis.2006.05.032. PMID 16806233.
- "Cloning, heterologous expression, and enzymological characterization of human squalene monooxygenase". Archives of Biochemistry and Biophysics 374 (2): 381–8. Feb 2000. doi:10.1006/abbi.1999.1629. PMID 10666321.
- "Squalene epoxidase, located on chromosome 8q24.1, is upregulated in 8q+ breast cancer and indicates poor clinical outcome in stage I and II disease". British Journal of Cancer 99 (5): 774–80. Sep 2008. doi:10.1038/sj.bjc.6604556. PMID 18728668.
- "SREBP-2 and NF-Y are involved in the transcriptional regulation of squalene epoxidase". Biochemical and Biophysical Research Communications 295 (1): 74–80. Jul 2002. doi:10.1016/S0006-291X(02)00623-X. PMID 12083769.
- "Identification of genes differentially expressed in human primary lung squamous cell carcinoma". Lung Cancer 56 (3): 307–17. Jun 2007. doi:10.1016/j.lungcan.2007.01.016. PMID 17316888.
- "Construction and characterization of a full length-enriched and a 5'-end-enriched cDNA library". Gene 200 (1–2): 149–56. Oct 1997. doi:10.1016/S0378-1119(97)00411-3. PMID 9373149.
- "Multiple genetic variants along candidate pathways influence plasma high-density lipoprotein cholesterol concentrations". Journal of Lipid Research 49 (12): 2582–9. Dec 2008. doi:10.1194/jlr.M800232-JLR200. PMID 18660489.
- "The LIFEdb database in 2006". Nucleic Acids Research 34 (Database issue): D415-8. Jan 2006. doi:10.1093/nar/gkj139. PMID 16381901.
- "DNA cloning using in vitro site-specific recombination". Genome Research 10 (11): 1788–95. Nov 2000. doi:10.1101/gr.143000. PMID 11076863.
- "Oligo-capping: a simple method to replace the cap structure of eukaryotic mRNAs with oligoribonucleotides". Gene 138 (1–2): 171–4. Jan 1994. doi:10.1016/0378-1119(94)90802-8. PMID 8125298.
- "Transcriptional regulation of squalene epoxidase by sterols and inhibitors in HeLa cells". The Journal of Biological Chemistry 271 (14): 8053–6. Apr 1996. doi:10.1074/jbc.271.14.8053. PMID 8626488.
- "Localization of the squalene epoxidase gene (SQLE) to human chromosome region 8q24.1". Genomics 44 (1): 141–3. Aug 1997. doi:10.1006/geno.1997.4825. PMID 9286711.
- "From ORFeome to biology: a functional genomics pipeline". Genome Research 14 (10B): 2136–44. Oct 2004. doi:10.1101/gr.2576704. PMID 15489336.
External links
- Squalene+monooxygenase at the US National Library of Medicine Medical Subject Headings (MeSH)
This article incorporates text from the United States National Library of Medicine, which is in the public domain.
 |
