Biology:Oncometabolism
Oncometabolism is the field of study that focuses on the metabolic changes that occur in cells that make up the tumor microenvironment (TME) and accompany oncogenesis and tumor progression toward a neoplastic state.[1] Cells with increased growth and survivability differ from non-tumorigenic cells in terms of metabolism.[2] The Warburg Effect, which describes how cancer cells change their metabolism to become more oncogenic in order to proliferate and eventually invade other tissues in a process known as metastasis.[1]
The chemical reactions associated with oncometabolism are triggered by the alteration of oncogenes, which are genes that have the potential to cause cancer.[3] These genes can be functional and active during physiological conditions, producing normal amounts of metabolites. Their upregulation as a result of DNA damage can result in an overabundance of these metabolites, and lead to tumorigenesis. These metabolites are known as oncometabolites, and can act as biomarkers.[4]
History
In the 1920s, Otto Heinrich Warburg discovered an intriguing bioenergetic phenotype shared by most tumor cells: a higher-than-normal reliance on lactic acid fermentation for energy generation. He is known as the "Father of Oncometabolism".[1][2] Although the roots of this research field trace back to the 1920s, it was only recently recognized[1] Over the last decade, research on cancer progression has focused on the role of shifting metabolic pathways for both the cancer and immune cells, leading to an increase interest in characterizing the metabolic alterations that cells undergo in the TME.[5]
Warburg Effect
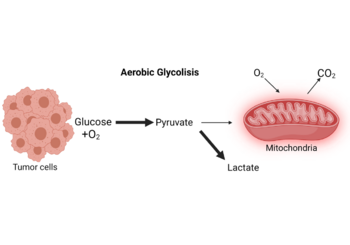
In the absence of hypoxic conditions (i.e. physiological levels of oxygen), cancer cells preferentially convert glucose to lactate, according to Otto H. Warburg, who believed that aerobic glycolysis was the key metabolic change in cancer cell malignancy. The "Warburg effect" was later coined to describe this metabolic shift.[6] Warburg thought this change in metabolism was due to mitochondrial "respiration injury", but this interpretation was questioned by other researchers in 1956 showing that intact and functional cytochromes detected in most tumor cells clearly speak against a general mitochondrial dysfunction.[7] Furthermore, Potter et al. and several other authors provided significant evidence that oxidative phosphorylation and a normal Krebs cycle persist in the vast majority malignant tumors, adding to the growing body of evidence that most cancers exhibit the Warburg effect while maintaining a proper mitochondrial respiration.[6][8] Dang et al.[9] in 2008 provided evidence that the tumor tissue sections used in Warburg's experiments should have been thinner for the oxygen diffusion constants employed, implying that the tissue slices studied were partially hypoxic and the calculated critical diffusion distance was of 470 micrometers.[6] As a result, endless debates and discussions about Warburg's discovery took place and have piqued the interest of scientists all over the world, which has helped bring attention to cell metabolism in cancer and immune cells and the use of modern technology to discover what these pathways are and how they are modified as well as potential therapeutic targets.
Metabolic reprogramming
Carcinogenic cells undergo a metabolic rewiring during oncogenesis, and oncometabolites play an important role. In cancer, there are several reprogrammed metabolic pathways that help cells survive when nutrients are scarce: Aerobic glycolysis, an increase in glycolytic flux, also known as the Warburg effect, allows glycolytic intermediates to supply subsidiary pathways to meet the metabolic demands of proliferating tumorigenic cells.[10] Another studied reprogrammed pathway is gain of function of the oncogene MYC. This gene encodes a transcription factor that boosts the expression of a number of genes involved in anabolic growth via mitochondrial metabolism.[11] Oncometabolite production is another example of metabolic deregulation.[12]
Oncometabolites
Oncometabolites are metabolites whose abundance increases markedly in cancer cells through loss-of-function or gain-of-function mutations in specific enzymes involved in their production, the accumulation of these endogenous metabolites initiates or sustains tumor growth and metastasis.[13] Cancer cells rely on aerobic glycolysis, which is reached through defects in enzymes involved in normal cell metabolism, this allows the cancer cells to meet their energy needs and divert acetyl-CoA from the TCA cycle to build essential biomolecules such as amino acids and lipids.[14] These defects cause an overabundance of endogenous metabolites, which are frequently involved in critical epigenetic changes and signaling pathways that have a direct impact on cancer cell metabolism.[15]
| Oncometabolite | Role | Oncogenes | Enzyme affected | Associated malignancies | References |
|---|---|---|---|---|---|
| D-2-hydroxyglutarate | Inhibits ATP synthase and mTOR signalling | IDH1
IDH2 |
Isocitrate dehydrogenase | Brain cancer, Leukemia | [15][16][17][13] |
| Succinate | Inhibits 2-oxoglutarate-dependent oxygenase | SDHA,SDHB,SDHC,SDHD,SDHAF1,SDHAF2 | succinate dehydrogenase | Renal and Thyroid tumors | [15][13] |
| Fumarate | Inhibits 2-oxoglutarate-dependent oxygenase | FH | Fumarate hydratase | Leiomyomata, Renal cysts | [15][16][13] |
| Glutamine* | *(Primary carbon-source for the biosynthesis of the oncometabolite 2-hydroxyglutarate) | [18][19] | |||
| Sarcosine | Activates mTOR signalling pathway | GNMT | Glycine-N-methyltransferase | Pancreatic cancer, Hepatocellular carcinoma | [18][20][21][13][22][23] |
| Asparagine | Anti-apoptotic agent | ASNS | Asparagine synthetase | Acute Lymphoblastic Leukemia | [18][13][24] |
| Choline | Methyl donor for DNA methylation which disrupts DNA repair | PCYT1A | phosphate cytidylyltransferase 1 choline-α | Breast, Brain and Prostate cancer | [18][13][24] |
| Lactate | Induces local immunosuppression | LDHA | Lactate dehydrogenase A | Various types of cancer[25] | [13][26] |
Epigenetics
Oncometabolite dysregulation and cancer progression are linked to epigenetic changes in cancer cells. Several mechanisms have been linked to D-2-hydroxyglutarate, succinate, and fumarate with the inhibition of α-KG–dependent dioxygenases, this causes epigenetic changes that affect the expression of genes involved in cell differentiation and the development of malignant characteristics.[27] The group of Timothy A. Chan[28] described a mechanism by which abnormal accumulation of the oncometabolite D-2-hydroxyglutarate in brain tumor samples increased DNA methylation, a process that has been shown to play a key role in oncogenesis.[29] On the other hand, in paraganglioma cells, succinate and fumarate were found to methylate histones, effectively silencing the genes PNMT and KRT19, which are involved in neuroendocrine differentiation and epithelial-mesenchymal transition, respectively.[30]
Biomarkers for cancer detection
The discovery of oncometabolites has ushered in a new era in cancer biology, one that has the potential to improve patient care. The discovery of new therapeutic and reliable markers that exploit vulnerabilities of cancer cells, are being used to targeting either upstream or downstream effectors of these pathways.[15] Oncometabolites can be used as diagnostic biomarkers and may be able to assist oncologists in making more precise decisions in early stages of tumorigenesis, particularly in predicting more aggressive tumor behavior.[4]
Isocitrate dehydrogenase
The detection of D-2-hydroxyglutarate in glioma patients using proton magnetic resonance spectroscopy (MRS) has been shown to be a noninvasive procedure. The presence of IDH1 or IDH2 mutations was linked to the detection of this oncometabolite 100 percent of the time.[31][27] IDH2/R140Q is a specific mutation that has shown promising results after its inhibition by the small molecule AGI-6780.[32] Therefore, limiting the supply of D-2-hydroxyglutarate by inhibiting the detected mutant IDH enzymes could be a good therapeutical approach to IDH-mutant cancers.[33]
Succinate dehydrogenase
IHC staining has been shown to be a useful diagnostic tool for prioritizing patients for SDH mutation testing in early stages of cancer. The absence of SDHB in IHC staining would be linked to the presence of SDH oncogene mutations.[34] The already commercialized drug decitabine (Dacogen®) could be an effective therapy to repress the migration capacities of SDHB-mutant cells,[30]
Fumarate hydratase
IHC staining for FH is used to detect lack of this protein in patients with papillary renal cell carcinoma type 2.[35] The lack of FH in renal carcinoma cells induces pro-survival metabolic adaptations where several cascades are affected.[36]
Glycine-N-methyltransferase
Downregulation of glycine-N-methyltransferase has been linked to hepatocellular carcinoma and pancreatic cancer. Serving this as a reliable marker for oncogenesis.[22] When compared to patients with deletions in GNMT, patients with no deletions early-stage pancreatic cancer had twice the median months overall survival.[23]
Applications
Oncometabolomics
Metabolomics can be applied to oncometabolism, since the changes in cancer's genomic, transcriptomic, and proteomic profiles can result in changes in downstream metabolic pathways. With this information we can elucidate the responsible pathways and oncometabolites for various diseases. Actually, through the use of this technique, the dysregulation of the pyruvate kinase enzyme in glucose metabolism was discovered in cancer cells. Another common used technique is glucose or glutamine labeled with 13C to show that the TCA cycle is used to generate large amounts of fatty acids (phospholipids) and to replenish the TCA cycle intermediates.[37] But oncometabolomics does not necessarily need to be used on cancer cells, but on cells immediately surrounding them in the TME.[38]
Metabolomics applied to cancer has the potential to significantly improve current oncological treatments and has a great diagnostic value, since metabolic changes are the prequel of phenotypic changes in cells (thus tissues and organs) making it suitable for early detection of difficult-to-detect cancers.[14] This also leads to a more personalized medicine and customize an individual's cancer treatment according to their specific oncometabolite profiles, which would allow for better cancer therapy customization or informed adjustments.[13][39]
Software and libraries
Ingenuity Pathway Analysis (IPA)
Ingenuity Pathway Analysis (IPA) is a metabolic pathway analysis software package that helps researchers model, analyze, and comprehend complex biological systems by associating specific metabolites with potential metabolic pathways for data analysis.[40] This software has been used by researchers to elucidate regulatory networks on oncometabolites like hydroxyglutarate.[41]
Metabolights
Metabolights is an open-access database for metabolomics research that collects all experimental data from leading journals' metabolic experiments.[42] Since its initial release in 2012, the MetaboLights repository has seen consistent year-on-year growth. It is a resource that surged in response to the needs of the scientific community to easy access to metabolite data.[43][44]
Research
Cancer research has been ongoing for centuries, trying to elucidate the origin of its cause. As cancer research evolves with time, the scientific community tends to pay more attention to cell metabolism and how to target these metabolic needs and changes that cells undergo during carcinogenesis.[45] There is growing evidence that metabolic dependencies in cancer are influenced by tissue environment, being this important to consider the TME for different in vitro and in vivo models to study oncometabolism in different cancer scenarios.[46]
There is extensive research on the modulation of BET proteins in cancer models of breast. These proteins appear to be involved in oncometabolism and targeting and uncoupling BRD4 actions in carcinogenic cells, as well as stopping pro-migratory signals and changing cytokine metabolism, particularly IL-6.[47] The same group has reported on the importance of exosomes in the TME and how these vesicles, shed by adipocytes, can carry a specific molecular cargo that causes metabolic changes in the cell, leading to pro-metastatic changes in the recipient breast cancer cells.[48]
References
- ↑ 1.0 1.1 1.2 1.3 "Otto Warburg: The journey towards the seminal discovery of tumor cell bioenergetic reprogramming". Biochimica et Biophysica Acta (BBA) - Molecular Basis of Disease 1867 (1): 165965. January 2021. doi:10.1016/j.bbadis.2020.165965. PMID 32949769.
- ↑ 2.0 2.1 ""Oncometabolism: The switchboard of cancer - An editorial"". Biochimica et Biophysica Acta (BBA) - Molecular Basis of Disease 1867 (2): 166031. February 2021. doi:10.1016/j.bbadis.2020.166031. PMID 33310398.
- ↑ "The Role of Oncogene Activation in Chemical Carcinogenesis". Chemical Carcinogenesis and Mutagenesis II. Handbook of Experimental Pharmacology. 94. Berlin, Heidelberg: Springer Berlin Heidelberg. 1990. pp. 319–352. doi:10.1007/978-3-642-74778-6_12. ISBN 978-3-642-74780-9.
- ↑ 4.0 4.1 "Oncometabolites in cancer aggressiveness and tumour repopulation". Biological Reviews of the Cambridge Philosophical Society 94 (4): 1530–1546. August 2019. doi:10.1111/brv.12513. PMID 30972955.
- ↑ "Metabolic Reprogramming of Immune Cells in Cancer Progression". Immunity 43 (3): 435–449. September 2015. doi:10.1016/j.immuni.2015.09.001. PMID 26377897.
- ↑ 6.0 6.1 6.2 "The Warburg effect: essential part of metabolic reprogramming and central contributor to cancer progression". International Journal of Radiation Biology 95 (7): 912–919. July 2019. doi:10.1080/09553002.2019.1589653. PMID 30822194.
- ↑ "The Warburg Effect 97 Years after Its Discovery". Cancers 12 (10): 2819. September 2020. doi:10.3390/cancers12102819. PMID 33008042.
- ↑ "The Warburg effect: 80 years on". Biochemical Society Transactions 44 (5): 1499–1505. October 2016. doi:10.1042/bst20160094. PMID 27911732.
- ↑ Dang, et al. The interplay between MYC and HIF in cancer. Nature Reviews Cancer volume 8, pages51–56 (2008).
- ↑ "Aerobic glycolysis: meeting the metabolic requirements of cell proliferation". Annual Review of Cell and Developmental Biology 27 (1): 441–464. 2011-11-10. doi:10.1146/annurev-cellbio-092910-154237. PMID 21985671.
- ↑ "Abstract IA05: Targeting MYC-mediated cancer metabolism". Molecular Cancer Research (American Association for Cancer Research) 13 (10_Supplement): IA05. October 2015. doi:10.1158/1557-3125.myc15-ia05.
- ↑ "Fundamentals of cancer metabolism". Science Advances 2 (5): e1600200. May 2016. doi:10.1126/sciadv.1600200. PMID 27386546. Bibcode: 2016SciA....2E0200D.
- ↑ 13.0 13.1 13.2 13.3 13.4 13.5 13.6 13.7 13.8 "Emerging applications of metabolomics in drug discovery and precision medicine". Nature Reviews. Drug Discovery 15 (7): 473–484. July 2016. doi:10.1038/nrd.2016.32. PMID 26965202.
- ↑ 14.0 14.1 "Oncometabolomics in cancer research". Expert Review of Proteomics 10 (4): 325–336. August 2013. doi:10.1586/14789450.2013.828947. PMID 23992416.
- ↑ 15.0 15.1 15.2 15.3 15.4 "Oncometabolites: A New Paradigm for Oncology, Metabolism, and the Clinical Laboratory". Clinical Chemistry 63 (12): 1812–1820. December 2017. doi:10.1373/clinchem.2016.267666. PMID 29038145.
- ↑ 16.0 16.1 "The emerging role of fumarate as an oncometabolite". Frontiers in Oncology 2: 85. 2012. doi:10.3389/fonc.2012.00085. PMID 22866264.
- ↑ "Oncometabolite? IDH1 discoveries raise possibility of new metabolism targets in brain cancers and leukemia". Journal of the National Cancer Institute 102 (13): 926–928. July 2010. doi:10.1093/jnci/djq262. PMID 20576929.
- ↑ 18.0 18.1 18.2 18.3 "Oncometabolites: A new insight for oncology". Molecular Genetics & Genomic Medicine 7 (9): e873. September 2019. doi:10.1002/mgg3.873. PMID 31321921.
- ↑ "In Vivo Imaging of Glutamine Metabolism to the Oncometabolite 2-Hydroxyglutarate in IDH1/2 Mutant Tumors". Cell Metabolism 26 (6): 830–841.e3. December 2017. doi:10.1016/j.cmet.2017.10.001. PMID 29056515.
- ↑ "The role of sarcosine metabolism in prostate cancer progression". Neoplasia 15 (5): 491–501. May 2013. doi:10.1593/neo.13314. PMID 23633921.
- ↑ "Sarcosine influences apoptosis and growth of prostate cells via cell-type specific regulation of distinct sets of genes". The Prostate 78 (2): 104–112. February 2018. doi:10.1002/pros.23450. PMID 29105933.
- ↑ 22.0 22.1 "Tumor suppressor gene glycine N-methyltransferase and its potential in liver disorders and hepatocellular carcinoma". Toxicology and Applied Pharmacology 378: 114607. September 2019. doi:10.1016/j.taap.2019.114607. PMID 31170416.
- ↑ 23.0 23.1 "The Case for GNMT as a Biomarker and a Therapeutic Target in Pancreatic Cancer". Pharmaceuticals 14 (3): 209. March 2021. doi:10.3390/ph14030209. PMID 33802396.
- ↑ 24.0 24.1 "Asparagine Synthetase in Cancer: Beyond Acute Lymphoblastic Leukemia". Frontiers in Oncology 9: 1480. 2020-01-09. doi:10.3389/fonc.2019.01480. PMID 31998641.
- ↑ "Lactate dehydrogenase A: A key player in carcinogenesis and potential target in cancer therapy". Cancer Medicine 7 (12): 6124–6136. December 2018. doi:10.1002/cam4.1820. PMID 30403008.
- ↑ "The Regulation and Function of Lactate Dehydrogenase A: Therapeutic Potential in Brain Tumor". Brain Pathology 26 (1): 3–17. January 2016. doi:10.1111/bpa.12299. PMID 26269128.
- ↑ 27.0 27.1 "Oncometabolites: linking altered metabolism with cancer". The Journal of Clinical Investigation 123 (9): 3652–3658. September 2013. doi:10.1172/jci67228. PMID 23999438.
- ↑ "IDH1 mutation is sufficient to establish the glioma hypermethylator phenotype". Nature 483 (7390): 479–483. February 2012. doi:10.1038/nature10866. PMID 22343889. Bibcode: 2012Natur.483..479T.
- ↑ "The epigenomics of cancer". Cell 128 (4): 683–692. February 2007. doi:10.1016/j.cell.2007.01.029. PMID 17320506.
- ↑ 30.0 30.1 "SDH mutations establish a hypermethylator phenotype in paraganglioma". Cancer Cell 23 (6): 739–752. June 2013. doi:10.1016/j.ccr.2013.04.018. PMID 23707781.
- ↑ "2-hydroxyglutarate detection by magnetic resonance spectroscopy in IDH-mutated patients with gliomas". Nature Medicine 18 (4): 624–629. January 2012. doi:10.1038/nm.2682. PMID 22281806.
- ↑ "Targeted inhibition of mutant IDH2 in leukemia cells induces cellular differentiation". Science 340 (6132): 622–626. May 2013. doi:10.1126/science.1234769. PMID 23558173. Bibcode: 2013Sci...340..622W.
- ↑ "Metabolism, Activity, and Targeting of D- and L-2-Hydroxyglutarates". Trends in Cancer 4 (2): 151–165. February 2018. doi:10.1016/j.trecan.2017.12.005. PMID 29458964.
- ↑ "An immunohistochemical procedure to detect patients with paraganglioma and phaeochromocytoma with germline SDHB, SDHC, or SDHD gene mutations: a retrospective and prospective analysis". The Lancet. Oncology 10 (8): 764–771. August 2009. doi:10.1016/S1470-2045(09)70164-0. PMID 19576851.
- ↑ "Fumarate Hydratase-deficient Renal Cell Carcinoma Is Strongly Correlated With Fumarate Hydratase Mutation and Hereditary Leiomyomatosis and Renal Cell Carcinoma Syndrome". The American Journal of Surgical Pathology 40 (7): 865–875. July 2016. doi:10.1097/pas.0000000000000617. PMID 26900816.
- ↑ "Fumarate hydratase in cancer: A multifaceted tumour suppressor". Seminars in Cell & Developmental Biology 98: 15–25. February 2020. doi:10.1016/j.semcdb.2019.05.002. PMID 31085323.
- ↑ "Global profiling strategies for mapping dysregulated metabolic pathways in cancer". Cell Metabolism 16 (5): 565–577. November 2012. doi:10.1016/j.cmet.2012.09.013. PMID 23063552.
- ↑ "Metabolic alterations in lung cancer-associated fibroblasts correlated with increased glycolytic metabolism of the tumor". Molecular Cancer Research 11 (6): 579–592. June 2013. doi:10.1158/1541-7786.mcr-12-0437-t. PMID 23475953.
- ↑ "Is Cancer a Genetic Disease or a Metabolic Disease?". eBioMedicine 2 (6): 478–479. June 2015. doi:10.1016/j.ebiom.2015.05.022. PMID 26288805.
- ↑ Analysis of metabolomic profiling data acquired on GC-MS. Methods in Enzymology. 543. Elsevier. 2014. pp. 315–324. doi:10.1016/B978-0-12-801329-8.00016-7. ISBN 9780128013298.
- ↑ "The oncometabolite R-2-hydroxyglutarate dysregulates the differentiation of human mesenchymal stromal cells via inducing DNA hypermethylation". BMC Cancer 21 (1): 36. January 2021. doi:10.1186/s12885-020-07744-x. PMID 33413208.
- ↑ "MetaboLights - Metabolomics experiments and derived information". https://www.ebi.ac.uk/metabolights/.
- ↑ "MetaboLights: a resource evolving in response to the needs of its scientific community". Nucleic Acids Research 48 (D1): D440–D444. January 2020. doi:10.1093/nar/gkz1019. PMID 31691833.
- ↑ "MetaboLights--an open-access general-purpose repository for metabolomics studies and associated meta-data". Nucleic Acids Research 41 (Database issue): D781–D786. January 2013. doi:10.1093/nar/gks1004. PMID 23109552.
- ↑ "Cancer Metabolism: Phenotype, Signaling and Therapeutic Targets". Cells 9 (10): 2308. October 2020. doi:10.3390/cells9102308. PMID 33081387.
- ↑ "Targeting Metabolism for Cancer Therapy". Cell Chemical Biology 24 (9): 1161–1180. September 2017. doi:10.1016/j.chembiol.2017.08.028. PMID 28938091.
- ↑ "BRD4 Regulates Breast Cancer Dissemination through Jagged1/Notch1 Signaling". Cancer Research 76 (22): 6555–6567. November 2016. doi:10.1158/0008-5472.CAN-16-0559. PMID 27651315.
- ↑ "Adipocyte-derived exosomes may promote breast cancer progression in type 2 diabetes". Science Signaling 14 (710): eabj2807. November 2021. doi:10.1126/scisignal.abj2807. ISSN 1945-0877. PMID 34813359.
 |