Biology:Phosphatidylethanolamine N-methyltransferase
| Phosphatidylethanolamine N-methyltransferase | |||||||||
|---|---|---|---|---|---|---|---|---|---|
| Identifiers | |||||||||
| EC number | 2.1.1.17 | ||||||||
| CAS number | 37256-91-0 | ||||||||
| Databases | |||||||||
| IntEnz | IntEnz view | ||||||||
| BRENDA | BRENDA entry | ||||||||
| ExPASy | NiceZyme view | ||||||||
| KEGG | KEGG entry | ||||||||
| MetaCyc | metabolic pathway | ||||||||
| PRIAM | profile | ||||||||
| PDB structures | RCSB PDB PDBe PDBsum | ||||||||
| Gene Ontology | AmiGO / QuickGO | ||||||||
| |||||||||
 Generic protein structure example |
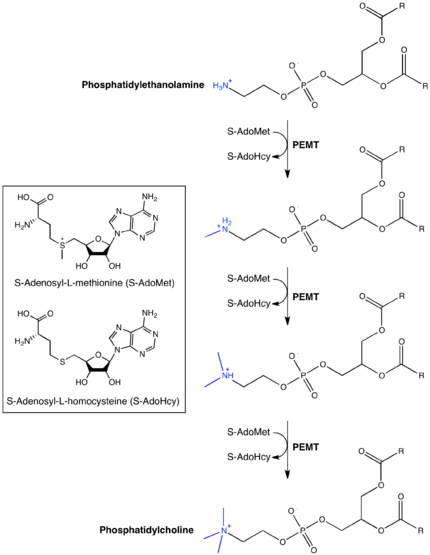
Phosphatidylethanolamine N-methyltransferase (abbreviated PEMT) is a transferase enzyme (EC 2.1.1.17) which converts phosphatidylethanolamine (PE) to phosphatidylcholine (PC) in the liver.[1][2][3] In humans it is encoded by the PEMT gene within the Smith–Magenis syndrome region on chromosome 17.[4][5]
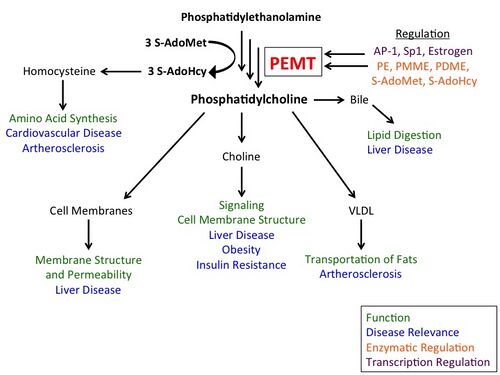
While the CDP-choline pathway, in which choline obtained either by dietary consumption or by metabolism of choline-containing lipids is converted to PC, accounts for approximately 70% of PC biosynthesis in the liver, the PEMT pathway has been shown to have played a critical evolutionary role in providing PC during times of starvation. Furthermore, PC made via PEMT plays a wide range of physiological roles, utilized in choline synthesis, hepatocyte membrane structure, bile secretion, and very low-density lipoprotein (VLDL) secretion.[6][7]
Nomenclature
Phosphatidylethanolamine N-methyltransferase is also known as lipid methyl transferase, LMTase, phosphatidylethanolamine methyltransferase, phosphatidylethanolamine-N-methylase, and phosphatidylethanolamine-S-adenosylmethionine-methyltransferase.
Function
The PEMT enzyme converts phosphatidylethanolamine (PE) to phosphatidylcholine (PC) via three sequential methylations by S-adenosyl methionine (SAM). The enzyme is found in endoplasmic reticulum and mitochondria-associated membranes. It accounts for ~30% of PC biosynthesis, with the CDP-choline, or Kennedy, pathway making ~70%.[6] PC, typically the most abundant phospholipid in animals and plants, accounts for more than half of cell membrane phospholipids and approximately 30% of all cellular lipid content. The PEMT pathway is therefore crucial for maintaining membrane integrity.[8]
PC made via the PEMT pathway can be degraded by phospholipases C/D, resulting in the de novo formation of choline. Thus, the PEMT pathway contributes to maintaining brain and liver function and larger-scale energy metabolism in the body.[3][6]
PC molecules produced by PEMT-catalyzed methylation of PE are more diverse, and tend to contain longer chain, polyunsaturated species and more arachidonate, whereas those produced via the CDP-choline pathway are typically composed of medium-length, saturated chains.[9]
A major pathway for hepatic PC utilization is secretion of bile into the intestine.[3] PEMT activity also dictates normal very low-density lipoprotein (VLDL) secretion by the liver.[10][11] PEMT is also a significant source and regulator of plasma homocysteine, which can be secreted or converted to methionine or cysteine.[12]
Mechanism
The exact mechanism by which PEMT catalyzes the sequential methylation of PE by three molecules of SAM to form PC remains unknown. Kinetic analyses as well as amino acid and gene sequencing have shed some light on how the enzyme works. Studies suggest that a single substrate binding site binds all three phospholipids methylated by PEMT: PE, phosphatidyl-monomethylethanolamine (PMME) and phosphatidyl-dimethylethanolamine. The first methylation, that of PE to PMME, has been shown to be the rate-limiting step in conversion of PE to PC. It is suspected that the structure or specific conformation adopted by PE has a lower affinity for the PEMT active site; consequently, upon methylation, PMME would be immediately converted to PDME and PDME to PC, via a Bi-Bi or ping-pong mechanism before another PE molecule could enter the active site.[3][13][14]
Structure
Purification of PEMT by Neale D. Ridgway and Dennis E. Vance in 1987 produced an 18.3 kDa protein.[15] Subsequent cloning, sequencing, and expression of PEMT cDNA resulted in a 22.3 kDa, 199-amino acid protein.[16] Although the enzymatic structure is unknown, PEMT is proposed to contain four hydrophobic membrane-spanning regions, with both its C and N termini on the cytosolic side of the ER membrane. Kinetic studies indicate a common binding site for PE, PMME, and PDME substrates.[3] SAM binding motifs have been identified on both the third and fourth transmembrane sequences. Site-directed mutagenesis has pinpointed the residues Gly98, Gly100, Glu180, and Glu181 to be essential for SAM binding in the active site.[17]
Regulation
PEMT activity is unrelated to enzyme mass, but rather is regulated by supply of substrates including PE, as well as PMME, PDME, and SAM. Low substrate levels inhibit PEMT. The enzyme is further regulated by S-adenosylhomocysteine produced after each methylation.[14][18][19]
PEMT gene expression is regulated by transcription factors including activator protein 1 (AP-1) and Sp1. Sp1 is a negative regulator of PEMT transcription, yet is it is a positive regulator of choline-phosphate cytidylyltransferase (CT) transcription.[3][20] This is one of several examples of the reciprocal regulation of PEMT and CT in the PEMT and CDP-choline pathways. Estrogen has also been shown to be a positive regulator of hepatocyte PEMT transcription. Ablation of the estrogen binding site in the PEMT promoter region may increase risk of hepatic steatosis from choline deficiency.[21]
Disease relevance
Liver
PEMT deficiency in mice, genetically induced by PEMT gene knockout, produced minimal effect on PE and PC levels. However, upon being fed a choline-deficient diet, the mice developed severe liver failure. Rapid PC depletion due to biliary PC secretion, as well as protein leakage from loss of membrane integrity due to lowered PC/PE ratios, led to steatosis and steatohepatitis.[6][22][23][24]
A Val-to-Met substitution at residue 175, leading to reduced PEMT activity, has been linked to non-alcoholic fatty liver disease.[25] This substitution has also been linked to increased frequency of non-alcoholic steatohepatitis.[26]
A single-nucleotide polymorphism (G to C) in the promoter region of the PEMT has been demonstrated to contribute to development of organ dysfunction in conjunction with a low-choline diet.[27]
Cardiovascular disease and atherosclerosis
PEMT modulates levels of blood plasma homocysteine, which is either secreted or converted to methionine or cysteine. High levels of homocysteine are linked to cardiovascular disease and atherosclerosis, particularly coronary artery disease.[28] PEMT deficiency prevents atherosclerosis in mice fed high-fat, high-cholesterol diets.[29] This is largely a result of lower levels of VLDL lipids in the PEMT-deficient mice.[30] Furthermore, the decreased lipid (PC) content in VLDLs causes changes in lipoprotein structure which allow them to be cleared more rapidly in the PEMT-deficient mice.[3]
Obesity and insulin resistance
PEMT-deficient mice fed high-fat diets have been shown to resist weight gain and be protected from insulin resistance. One potential reason for this phenomenon is that these mice, which exhibit hypermetabolic behavior, rely more on glucose than on fats for energy.[31] It was concluded that insufficient choline resulted in the lack of weight gain, supported by the fact that PC produced via the PEMT pathway can be used to form choline.[32]
The PEMT deficient mice showed elevated plasma glucagon levels, increased hepatic expression of glucagon receptor, phosphorylated AMP-activated protein kinase (AMPK), and serine-307-phosphorylated insulin receptor substrate 1 (IRS1-s307), which blocks insulin-mediated signal transduction; together, these contribute to enhanced gluconeogenesis and ultimately insulin resistance.[33] Another possibility is that lack of PEMT in adipose tissue may affect normal fat deposition.[34]
See also
References
- ↑ "Hepatic phosphatidylethanolamine N-methyltransferase, unexpected roles in animal biochemistry and physiology". The Journal of Biological Chemistry 282 (46): 33237–41. Nov 2007. doi:10.1074/jbc.R700028200. PMID 17881348.
- ↑ "EC 2.1.1.17". School of Biological and Chemical Sciences, Queen Mary, University of London. 17 February 2014. http://www.chem.qmul.ac.uk/iubmb/enzyme/EC2/1/1/17.html.
- ↑ 3.0 3.1 3.2 3.3 3.4 3.5 3.6 "Physiological roles of phosphatidylethanolamine N-methyltransferase". Biochimica et Biophysica Acta (BBA) - Molecular and Cell Biology of Lipids 1831 (3): 626–32. Mar 2013. doi:10.1016/j.bbalip.2012.07.017. PMID 22877991. https://zenodo.org/record/895264.
- ↑ "Entrez Gene: PEMT". https://www.ncbi.nlm.nih.gov/sites/entrez?Db=gene&Cmd=ShowDetailView&TermToSearch=10400.
- ↑ "Identification of three novel cDNAs for human phosphatidylethanolamine N-methyltransferase and localization of the human gene on chromosome 17p11.2". Biochimica et Biophysica Acta (BBA) - Molecular and Cell Biology of Lipids 1436 (3): 405–12. Jan 1999. doi:10.1016/s0005-2760(98)00147-7. PMID 9989271.
- ↑ 6.0 6.1 6.2 6.3 "Phospholipid methylation in mammals: from biochemistry to physiological function". Biochimica et Biophysica Acta (BBA) - Biomembranes 1838 (6): 1477–87. Jun 2014. doi:10.1016/j.bbamem.2013.10.018. PMID 24184426.
- ↑ "CTP: Phosphocholine cytidylyltransferase: paving the way from gene to membrane". The Journal of Biological Chemistry 280 (2): 853–6. Jan 2005. doi:10.1074/jbc.R400031200. PMID 15536089.
- ↑ Christie, William W., ed (16 September 2013). "Phosphatidylcholine and Related Lipids". AOCS. http://lipidlibrary.aocs.org/Lipids/pc/index.htm.
- ↑ "Molecular distinction of phosphatidylcholine synthesis between the CDP-choline pathway and phosphatidylethanolamine methylation pathway". The Journal of Biological Chemistry 274 (42): 29683–8. Oct 1999. doi:10.1074/jbc.274.42.29683. PMID 10514439.
- ↑ "The active synthesis of phosphatidylcholine is required for very low density lipoprotein secretion from rat hepatocytes". The Journal of Biological Chemistry 263 (6): 2998–3004. Feb 1988. doi:10.1016/S0021-9258(18)69166-5. PMID 3343237.
- ↑ "The role of phosphatidylcholine biosynthesis in the secretion of lipoproteins from hepatocytes". Canadian Journal of Biochemistry and Cell Biology 63 (8): 870–81. Aug 1985. doi:10.1139/o85-108. PMID 3904950.
- ↑ "Homocysteine and cardiovascular disease". Annual Review of Medicine 49: 31–62. 1998. doi:10.1146/annurev.med.49.1.31. PMID 9509248.
- ↑ "Kinetic mechanism of phosphatidylethanolamine N-methyltransferase". The Journal of Biological Chemistry 263 (32): 16864–71. Nov 1988. doi:10.1016/S0021-9258(18)37471-4. PMID 3182819.
- ↑ 14.0 14.1 "Phosphatidylethanolamine levels and regulation of phosphatidylethanolamine N-methyltransferase". The Journal of Biological Chemistry 264 (2): 1203–7. Jan 1989. doi:10.1016/S0021-9258(19)85072-X. PMID 2910850.
- ↑ "Purification of phosphatidylethanolamine N-methyltransferase from rat liver". The Journal of Biological Chemistry 262 (35): 17231–9. Dec 1987. doi:10.1016/S0021-9258(18)45514-7. PMID 3680298.
- ↑ "Cloning and expression of a novel phosphatidylethanolamine N-methyltransferase. A specific biochemical and cytological marker for a unique membrane fraction in rat liver". The Journal of Biological Chemistry 268 (22): 16655–63. Aug 1993. doi:10.1016/S0021-9258(19)85468-6. PMID 8344945.
- ↑ "Molecular dissection of the S-adenosylmethionine-binding site of phosphatidylethanolamine N-methyltransferase". The Journal of Biological Chemistry 278 (37): 35826–36. Sep 2003. doi:10.1074/jbc.M306308200. PMID 12842883.
- ↑ "Regulation of phospholipid biosynthesis in isolated rat hepatocytes. Effect of different substrates". The Journal of Biological Chemistry 250 (9): 3359–67. May 1975. doi:10.1016/S0021-9258(19)41523-8. PMID 1123345.
- ↑ "The methylation of phosphatidylethanolamine". Progress in Lipid Research 27 (1): 61–79. 1988. doi:10.1016/0163-7827(88)90005-7. PMID 3057511.
- ↑ "A role for Sp1 in transcriptional regulation of phosphatidylethanolamine N-methyltransferase in liver and 3T3-L1 adipocytes". The Journal of Biological Chemistry 285 (16): 11880–91. Apr 2010. doi:10.1074/jbc.M110.109843. PMID 20150657.
- ↑ "Aberrant estrogen regulation of PEMT results in choline deficiency-associated liver dysfunction". The Journal of Biological Chemistry 286 (2): 1649–58. Jan 2011. doi:10.1074/jbc.M110.106922. PMID 21059658.
- ↑ "Biochemical and evolutionary significance of phospholipid methylation". The Journal of Biological Chemistry 273 (42): 27043–6. Oct 1998. doi:10.1074/jbc.273.42.27043. PMID 9765216.
- ↑ "Homozygous disruption of the murine mdr2 P-glycoprotein gene leads to a complete absence of phospholipid from bile and to liver disease". Cell 75 (3): 451–62. Nov 1993. doi:10.1016/0092-8674(93)90380-9. PMID 8106172.
- ↑ "The ratio of phosphatidylcholine to phosphatidylethanolamine influences membrane integrity and steatohepatitis". Cell Metabolism 3 (5): 321–31. May 2006. doi:10.1016/j.cmet.2006.03.007. PMID 16679290.
- ↑ "Polymorphism of the PEMT gene and susceptibility to nonalcoholic fatty liver disease (NAFLD)". FASEB Journal 19 (10): 1266–71. Aug 2005. doi:10.1096/fj.04-3580com. PMID 16051693.
- ↑ Zeisel, S. H. (2006). "People with fatty liver are more likely to have the PEMT rs7946 SNP, yet populations with the mutant allele do not have fatty liver". The FASEB Journal 20 (12): 2181–2182. doi:10.1096/fj.06-1005ufm.
- ↑ "Common genetic polymorphisms affect the human requirement for the nutrient choline". FASEB Journal 20 (9): 1336–44. Jul 2006. doi:10.1096/fj.06-5734com. PMID 16816108.
- ↑ Robinson, Killian H. (2001). "Homocysteine and coronary artery disease". in Carmel, Ralph; Jacobsen, Ralph Carmel. Homocysteine in Health and Disease. Cambridge: Cambridge University Press. pp. 371–383.
- ↑ "Lack of phosphatidylethanolamine N-methyltransferase alters plasma VLDL phospholipids and attenuates atherosclerosis in mice". Arteriosclerosis, Thrombosis, and Vascular Biology 29 (9): 1349–55. Sep 2009. doi:10.1161/ATVBAHA.109.188672. PMID 19520976.
- ↑ "An unexpected requirement for phosphatidylethanolamine N-methyltransferase in the secretion of very low density lipoproteins". The Journal of Biological Chemistry 277 (44): 42358–65. Nov 2002. doi:10.1074/jbc.M204542200. PMID 12193594.
- ↑ "Impaired de novo choline synthesis explains why phosphatidylethanolamine N-methyltransferase-deficient mice are protected from diet-induced obesity". The Journal of Biological Chemistry 285 (29): 22403–13. Jul 2010. doi:10.1074/jbc.M110.108514. PMID 20452975.
- ↑ Zeisel, Steven H. (1987). "Phosphatidylcholine: Endogenous Precursor of Choline". in Hanin, Israel; Ansell, Gordon Brian. Lecithin: Technological, Biological and Therapeutic Aspects. New York: Plenum Press. pp. 107–120.
- ↑ "Choline supplementation promotes hepatic insulin resistance in phosphatidylethanolamine N-methyltransferase-deficient mice via increased glucagon action". The Journal of Biological Chemistry 288 (2): 837–47. Jan 2013. doi:10.1074/jbc.M112.415117. PMID 23179947.
- ↑ "Sequential synthesis and methylation of phosphatidylethanolamine promote lipid droplet biosynthesis and stability in tissue culture and in vivo". The Journal of Biological Chemistry 286 (19): 17338–50. May 2011. doi:10.1074/jbc.M111.234534. PMID 21454708.
Further reading
- "Identification and properties of two methyltransferases in conversion of phosphatidylethanolamine to phosphatidylcholine". Proceedings of the National Academy of Sciences of the United States of America 75 (4): 1718–21. Apr 1978. doi:10.1073/pnas.75.4.1718. PMID 25437. Bibcode: 1978PNAS...75.1718H.
- "Isolation and characterization of lipid N-methylrtansferase from dog lung". Biochimica et Biophysica Acta (BBA) - Enzymology 178 (1): 21–34. Mar 1969. doi:10.1016/0005-2744(69)90128-4. PMID 5773456.
- "Conversion of phosphatidylethanolamine to phosphatidylcholine in rat liver. Partial purification and characterization of the enzymatic activities". The Journal of Biological Chemistry 254 (10): 3886–91. May 1979. doi:10.1016/S0021-9258(18)50670-0. PMID 438165.
- "Genome-wide association study of biochemical traits in Korcula Island, Croatia". Croatian Medical Journal 50 (1): 23–33. Feb 2009. doi:10.3325/cmj.2009.50.23. PMID 19260141.
- "Associations of folate and choline metabolism gene polymorphisms with orofacial clefts". Journal of Medical Genetics 47 (12): 809–15. Dec 2010. doi:10.1136/jmg.2009.070029. PMID 19737740.
- "Polymorphism of the PEMT gene and susceptibility to nonalcoholic fatty liver disease (NAFLD)". FASEB Journal 19 (10): 1266–71. Aug 2005. doi:10.1096/fj.04-3580com. PMID 16051693.
- "Genetic variants in phosphatidylethanolamine N-methyltransferase and methylenetetrahydrofolate dehydrogenase influence biomarkers of choline metabolism when folate intake is restricted". Journal of the American Dietetic Association 109 (2): 313–8. Feb 2009. doi:10.1016/j.jada.2008.10.046. PMID 19167960.
- "Common genetic polymorphisms affect the human requirement for the nutrient choline". FASEB Journal 20 (9): 1336–44. Jul 2006. doi:10.1096/fj.06-5734com. PMID 16816108.
- "Association study between single-nucleotide polymorphisms in 199 drug-related genes and commonly measured quantitative traits of 752 healthy Japanese subjects". Journal of Human Genetics 54 (6): 317–23. Jun 2009. doi:10.1038/jhg.2009.31. PMID 19343046.
- "Phosphatidylethanolamine N-methyltransferase from liver". Biochimica et Biophysica Acta (BBA) - Lipids and Lipid Metabolism 1348 (1–2): 142–50. Sep 1997. doi:10.1016/s0005-2760(97)00108-2. PMID 9370326.
- "The phosphatidylethanolamine N-methyltransferase gene V175M single nucleotide polymorphism confers the susceptibility to NASH in Japanese population". Journal of Hepatology 46 (5): 915–20. May 2007. doi:10.1016/j.jhep.2006.12.012. PMID 17391797.
- "Phosphatidylethanolamine N-methyltransferase (PEMT) gene expression is induced by estrogen in human and mouse primary hepatocytes". FASEB Journal 21 (10): 2622–32. Aug 2007. doi:10.1096/fj.07-8227com. PMID 17456783.
- "No association of the rs4646396 SNP in the PEMT locus with schizophrenia in a Chinese case-control sample". Psychiatry Research 169 (2): 176–7. Sep 2009. doi:10.1016/j.psychres.2008.11.004. PMID 19647326.
- "Choline intake, plasma riboflavin, and the phosphatidylethanolamine N-methyltransferase G5465A genotype predict plasma homocysteine in folate-deplete Mexican-American men with the methylenetetrahydrofolate reductase 677TT genotype". The Journal of Nutrition 139 (4): 727–33. Apr 2009. doi:10.3945/jn.108.100222. PMID 19211833.
- "Localization-independent regulation of homocysteine secretion by phosphatidylethanolamine N-methyltransferase". The Journal of Biological Chemistry 280 (29): 27339–44. Jul 2005. doi:10.1074/jbc.M504658200. PMID 15927961.
- "A study of the PEMT gene in schizophrenia". Neuroscience Letters 424 (3): 203–6. Sep 2007. doi:10.1016/j.neulet.2007.07.038. PMID 17720317.
- "Molecular dissection of the S-adenosylmethionine-binding site of phosphatidylethanolamine N-methyltransferase". The Journal of Biological Chemistry 278 (37): 35826–36. Sep 2003. doi:10.1074/jbc.M306308200. PMID 12842883.
- "Choline metabolism and risk of breast cancer in a population-based study". FASEB Journal 22 (6): 2045–52. Jun 2008. doi:10.1096/fj.07-101279. PMID 18230680.
- "Expression of phosphatidylethanolamine N-methyltransferase in human hepatocellular carcinomas". Oncology 65 (2): 152–8. 2003. doi:10.1159/000072341. PMID 12931022.
- "Polymorphisms of microsomal triglyceride transfer protein gene and phosphatidylethanolamine N-methyltransferase gene in alcoholic and nonalcoholic fatty liver disease in Koreans". European Journal of Gastroenterology & Hepatology 21 (6): 667–72. Jun 2009. doi:10.1097/MEG.0b013e3283196adc. PMID 19262398.
- "Candidate genetic analysis of plasma high-density lipoprotein-cholesterol and severity of coronary atherosclerosis". BMC Medical Genetics 10: 111. 2009. doi:10.1186/1471-2350-10-111. PMID 19878569.
External links
- Phosphatidylethanolamine+N-Methyltransferase at the US National Library of Medicine Medical Subject Headings (MeSH)
This article incorporates text from the United States National Library of Medicine, which is in the public domain.
 |