Biology:Phospholipase D
| Phospholipase D | |||||||||
|---|---|---|---|---|---|---|---|---|---|
| Identifiers | |||||||||
| Symbol | PLDc | ||||||||
| Pfam | PF03009 | ||||||||
| InterPro | IPR001736 | ||||||||
| SMART | SM00155 | ||||||||
| PROSITE | PDOC50035 | ||||||||
| SCOP2 | 1byr / SCOPe / SUPFAM | ||||||||
| OPM superfamily | 118 | ||||||||
| OPM protein | 3rlh | ||||||||
| CDD | cd00138 | ||||||||
| Membranome | 306 | ||||||||
| |||||||||
| phospholipase D | |||||||||
|---|---|---|---|---|---|---|---|---|---|
| Identifiers | |||||||||
| EC number | 3.1.4.4 | ||||||||
| CAS number | 9001-87-0 | ||||||||
| Databases | |||||||||
| IntEnz | IntEnz view | ||||||||
| BRENDA | BRENDA entry | ||||||||
| ExPASy | NiceZyme view | ||||||||
| KEGG | KEGG entry | ||||||||
| MetaCyc | metabolic pathway | ||||||||
| PRIAM | profile | ||||||||
| PDB structures | RCSB PDB PDBe PDBsum | ||||||||
| Gene Ontology | AmiGO / QuickGO | ||||||||
| |||||||||
Phospholipase D (EC 3.1.4.4, lipophosphodiesterase II, lecithinase D, choline phosphatase, PLD; systematic name phosphatidylcholine phosphatidohydrolase) is an enzyme of the phospholipase superfamily that catalyses the following reaction
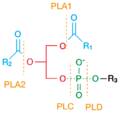
- a phosphatidylcholine + H2O = choline + a phosphatidate
Phospholipases occur widely, and can be found in a wide range of organisms, including bacteria, yeast, plants, animals, and viruses.[1][2] Phospholipase D's principal substrate is phosphatidylcholine, which it hydrolyzes to produce the signal molecule phosphatidic acid (PA), and soluble choline in a cholesterol dependent process called substrate presentation.[3] Plants contain numerous genes that encode various PLD isoenzymes, with molecular weights ranging from 90 to 125 kDa.[4] Mammalian cells encode two isoforms of phospholipase D: PLD1 and PLD2.[5] Phospholipase D is an important player in many physiological processes, including membrane trafficking, cytoskeletal reorganization, receptor-mediated endocytosis, exocytosis, and cell migration.[6] Through these processes, it has been further implicated in the pathophysiology of multiple diseases: in particular the progression of Parkinson's and Alzheimer's, as well as various cancers.[4][6] PLD may also help set the threshold for sensitivity to anesthesia and mechanical force.[7][8]
Discovery
PLD-type activity was first reported in 1947 by Donald J. Hanahan and I.L. Chaikoff.[1] It was not until 1975, however, that the hydrolytic mechanism of action was elucidated in mammalian cells. Plant isoforms of PLD were first purified from cabbage and castor bean; PLDα was ultimately cloned and characterized from a variety of plants, including rice, corn, and tomato.[1] Plant PLDs have been cloned in three isoforms: PLDα, PLDβ, and PLDγ.[9] More than half a century of biochemical studies have implicated phospholipase D and PA activity in a wide range of physiological processes and diseases, including inflammation, diabetes, phagocytosis, neuronal & cardiac signaling, and oncogenesis.[10]
Function
Strictly speaking, phospholipase D is a transphosphatidylase: it mediates the exchange of polar headgroups covalently attached to membrane-bound lipids. Utilizing water as a nucleophile, this enzyme catalyzes the cleavage of the phosphodiester bond in structural phospholipids such as phosphatidylcholine and phosphatidylethanolamine.[4] The products of this hydrolysis are the membrane-bound lipid phosphatidic acid (PA), and choline, which diffuses into the cytosol. As choline has little second messenger activity, PLD activity is mostly transduced by the production of PA.[6][11] PA is heavily involved in intracellular signal transduction.[12] In addition, some members of the PLD superfamily may employ primary alcohols such as ethanol or 1-butanol in the cleavage of the phospholipid, effectively catalyzing the exchange the polar lipid headgroup.[4][9] Other members of this family are able hydrolyze other phospholipid substrates, such as cardiolipin, or even the phosphodiester bond constituting the backbone of DNA.[5]
Phosphatidic acid
Many of phospholipase D's cellular functions are mediated by its principal product, phosphatidic acid (PA). PA is a negatively charged phospholipid, whose small head group promotes membrane curvature.[5] It is thus thought to facilitate membrane-vesicle fusion and fission in a manner analogous to clathrin-mediated endocytosis.[5] PA may also recruit proteins that contain its corresponding binding domain, a region characterized by basic amino acid-rich regions. Additionally, PA can be converted into a number of other lipids, such as lysophosphatidic acid (lyso-PA) or diacylglycerol, signal molecules which have a multitude of effects on downstream cellular pathways.[9] PA and its lipid derivatives are implicated in myriad processes that include intracellular vesicle trafficking, endocytosis, exocytosis, actin cytoskeleton dynamics, cell proliferation differentiation, and migration.[5]
Mammalian PLD directly interacts with kinases like PKC, ERK, TYK and controls the signalling indicating that PLD is activated by these kinases.[13] As choline is very abundant in the cell, PLD activity does not significantly affect choline levels, and choline is unlikely to play any role in signalling.
Phosphatidic acid is a signal molecule and acts to recruit SK1 to membranes. PA is extremely short lived and is rapidly hydrolysed by the enzyme phosphatidate phosphatase to form diacylglycerol (DAG). DAG may also be converted to PA by DAG kinase. Although PA and DAG are interconvertible, they do not act in the same pathways. Stimuli that activate PLD do not activate enzymes downstream of DAG and vice versa.
It is possible that, though PA and DAG are interconvertible, separate pools of signalling and non-signalling lipids may be maintained. Studies have suggested that DAG signalling is mediated by polyunsaturated DAG while PLD derived PA is monounsaturated or saturated. Thus functional saturated/monounsaturated PA can be degraded by hydrolysing it to form non-functional saturated/monounsaturated DAG while functional polyunsaturated DAG can be degraded by converting it into non-functional polyunsaturated PA.[14][15][16]
A lysophospholipase D called autotaxin was recently identified as having an important role in cell-proliferation through its product, lysophosphatidic acid (LPA).
Structure
Plant and animal PLDs have a consistent molecular structure, characterized by sites of catalysis surrounded by an assortment of regulatory sequences.[4] The active site of PLDs consists of four highly conserved amino acid sequences (I-IV), of which motifs II and IV are particularly conserved. These structural domains contain the distinguishing catalytic sequence HxKxxxxD (HKD), where H, K, and D are the amino acids histidine (H), lysine (K), aspartic acid (D), while x represents nonconservative amino acids.[4][5] These two HKD motifs confer hydrolytic activity to PLD, and are critical for its enzymatic activity both in vitro and in vivo.[5][10] Hydrolysis of the phosphodiester bond occurs when these HKD sequences are in the correct proximity.
Human proteins containing this motif include:
PC-hydrolyzing PLD is a homologue of cardiolipin synthase,[17][18] phosphatidylserine synthase, bacterial PLDs, and viral proteins. Each of these appears to possess a domain duplication which is apparent by the presence of two HKD motifs containing well-conserved histidine, lysine, and asparagine residues which may contribute to the active site aspartic acid. An Escherichia coli endonuclease (nuc) and similar proteins appear to be PLD homologues but possess only one of these motifs.[19][20][21][22]
PLD genes additionally encode highly conserved regulatory domains: the phox consensus sequence (PX), the pleckstrin homology domain (PH), and a binding site for phosphatidylinositol 4,5-bisphosphate (PIP2).[2]
Mechanism of catalysis
PLD-catalyzed hydrolysis has been proposed to occur in two stages via a "ping-pong" mechanism. In this scheme, the histidine residues of each HKD motif successively attack the phospholipid substrate. Functioning as nucleophiles, the constituent imidazole moieties of the histidines form transient covalent bonds with the phospholipid, producing a short-lived intermediate that can be easily hydrolyzed by water in a subsequent step.[4][12]

Mechanism of activation
Substrate presentation For mammalian PLD2, the molecular basis of activation is substrate presentation. The enzyme resides inactive in lipid micro-domains rich in sphingomyelin and depleted of PC substrate.[23] Increased PIP2 or a decrease in cholesterol causes the enzyme to translocate to PIP2 micro domains near its substrate PC. Hence PLD can is primarily activated by localization within the plasma membrane rather than a protein conformational change. Disruption of lipid domains by anesthetics.[24] or mechanical force.[23] The protein may also undergo a conformational change upon PIP2 binding, but this has not been shown experimentally and would constitute a mechanism of activation distinct from substrate presentation.
Isoforms
Two major isoforms of phospholipase D has been identified in mammalian cells: PLD1 and PLD2 (53% sequence homology),[25] each encoded by distinct genes.[5] PLD activity appears to be present in most cell types, with the possible exceptions of peripheral leukocytes and other lymphocytes.[10] Both PLD isoforms require PIP2 as a cofactor for activity.[5] PLD1 and PLD2 exhibit different subcellular localizations that dynamically change in the course of signal transduction. PLD activity has been observed within the plasma membrane, cytosol, ER, and Golgi complex.[10]
PLD1
PLD1 is a 120 kDa protein that is mainly located on the inner membranes of cells. It is primarily present at the Golgi complex, endosomes, lysosomes, and secretory granules.[5] Upon the binding of an extracellular stimulus, PLD1 is transported to the plasma membrane. Basal PLD1 activity is low however, and in order to transduce the extracellular signal, it must first be activated by proteins such as Arf, Rho, Rac, and protein kinase C.[5][6][11]
PLD2In contrast, PLD2 is a 106 kDa protein that primarily localizes to the plasma membrane, residing in light membrane lipid rafts.[4][6] It has high intrinsic catalytic activity, and is only weakly activated by the above molecules.[4] |
| ||||||||||||||||||||||||||||||||||||||||||||
Regulation
The activity of phospholipase D is extensively regulated by hormones, neurotransmitters, lipids, small monomeric GTPases, and other small molecules that bind to their corresponding domains on the enzyme.[4] In most cases, signal transduction is mediated through production of phosphatidic acid, which functions as a secondary messenger.[4]
Specific phospholipids are regulators of PLD activity in plant and animal cells.[1][4] Most PLDs require phosphatidylinositol 4,5-bisphosphate (PIP2), as a cofactors for activity.[2][4] PIP2 and other phosphoinositides are important modifiers of cytoskeletal dynamics and membrane transport and can traffic PLD to its substrate PC.[26] PLDs regulated by these phospholipids are commonly involved in intracellular signal transduction.[4] Their activity is dependent upon the binding of these phosphoinositides near the active site.[4] In plants and animals, this binding site is characterized by the presence of a conserved sequence of basic and aromatic amino acids.[4][12] In plants such as Arabidopsis thaliana, this sequence is constituted by a RxxxxxKxR motif together with its inverted repeat, where R is arginine and K is lysine. Its proximity to the active site ensures high level of PLD1 and PLD2 activity, and promotes the translocation of PLD1 to target membranes in response to extracellular signals.[4]
C2 domain
Calcium acts as a cofactor in PLD isoforms that contain the C2 domain. Binding of Ca2+ to the C2 domain leads to conformational changes in the enzyme that strengthen enzyme-substrate binding, while weakening the association with phosphoinositides. In some plant isoenzymes, such as PLDβ, Ca2+ may bind directly to the active site, indirectly increasing its affinity for the substrate by strengthening the binding of the activator PIP2.[4]
PX domain
The pbox consensus sequence (PX) is thought to mediate the binding of additional phosphatidylinositol phosphates, in particular, phosphatidylinositol 5-phosphate (PtdIns5P), a lipid thought to be required for endocytosis, may help facilitate the reinternalization of PLD1 from the plasma membrane.[1]
PH domain
The highly conserved Pleckstrin homology domain (PH) is a structural domain approximately 120 amino acids in length. It binds phosphatidylinositides such as phosphatidylinositol (3,4,5)-trisphosphate (PIP3) and phosphatidylinositol (4,5)-bisphosphate (PIP2). It may also bind heterotrimeric G proteins via their βγ-subunit. Binding to this domain is also thought to facilitate the re-internalization of the protein by increasing its affinity to endocytotic lipid rafts.[1]
Interactions with small GTPases
In animal cells, small protein factors are important additional regulators of PLD activity. These small monomeric GTPases are members of the Rho and ARF families of the Ras superfamily. Some of these proteins, such as Rac1, Cdc42, and RhoA, allosterically activate mammalian PLD1, directly increasing its activity. In particular, the translocation of cytosolic ADP-ribosylation factor (ARF) to the plasma membrane is essential for PLD activation.[1][4]
Physiological and pathophysiological roles
Alcohol Intoxication
Phospholipase D metabolizes ethanol into phosphatidylethanol (PEtOH) in a process termed transphosphatidylation. Using fly genetics the PEtOH was shown to mediates alcohol's hyperactive response in fruit flies.[27] And ethanol transphosphatidylation was shown to be up-regulated in alcoholics and the family members of alcoholic.s[28] This ethanol transphosphatidylation mechanism recently emerged as an alternative theory for alcohol's effect on ion channels. Many ion channels are regulated by anionic lipids.[29] and the competition of PEtOH with endogenous signaling lipids is thought to mediate the effect of ethanol on ion channels in some instances and not direct binding of the free ethanol to the channel.[27]
Mechanosensation
PLD2 is a mechanosensor and directly sensitive to mechanical disruption of clustered GM1 lipids.[3] Mechanical disruption (fluid shear) then signals for the cell to differentiate. PLD2 also activates TREK-1 channels, a potassium channel in the analgesic pathway.[30]
In cancer
Phospholipase D is a regulator of several critical cellular processes, including vesicle transport, endocytosis, exocytosis, cell migration, and mitosis.[6] Dysregulation of these processes is commonplace in carcinogenesis,[6] and in turn, abnormalities in PLD expression have been implicated in the progression of several types cancer.[2][5] A driver mutation conferring elevated PLD2 activity has been observed in several malignant breast cancers.[5] Elevated PLD expression has also been correlated with tumor size in colorectal carcinoma, gastric carcinoma, and renal cancer.[5][6] However, the molecular pathways through which PLD drives cancer progression remain unclear.[5] One potential hypothesis casts a critical role for phospholipase D in the activation of mTOR, a suppressor of cancer cell apoptosis.[5] The ability of PLD to suppress apoptosis in cells with elevated tyrosine kinase activity makes it a candidate oncogene in cancers where such expression is typical.[6]
In neurodegenerative diseases
Phospholipase D may also play an important pathophysiological role in the progression of neurodegenerative diseases, primarily through its capacity as a signal transducer in indispensable cellular processes like cytoskeletal reorganization and vesicle trafficking.[25] Dysregulation of PLD by the protein α-synuclein has been shown to lead to the specific loss of dopaminergic neurons in mammals. α-synuclein is the primary structural component of Lewy bodies, protein aggregates that are the hallmarks of Parkinson's disease.[5] Disinhibition of PLD by α-synuclein may contribute to Parkinson's deleterious phenotype.[5]
Abnormal PLD activity has also been suspected in Alzheimer's disease, where it has been observed to interact with presenilin 1 (PS-1), the principal component of the γ-secretase complex responsible for the enzymatic cleavage of amyloid precursor protein (APP). Extracellular plaques of the product β-amyloid are a defining feature of Alzheimer's diseased brains.[5] Action of PLD1 on PS-1 has been shown to affect the intracellular trafficking of the amyloid precursor to this complex.[5][25] Phospholipase D3 (PLD3), a non-classical and poorly characterized member of the PLD superfamily, has also been associated with the pathogenesis of this disease.[31]
Gallery
References
- ↑ 1.0 1.1 1.2 1.3 1.4 1.5 1.6 "Phospholipase D: a lipid centric review". Cellular and Molecular Life Sciences 62 (19–20): 2305–16. October 2005. doi:10.1007/s00018-005-5195-z. PMID 16143829.
- ↑ 2.0 2.1 2.2 2.3 "Phospholipase D-structure, regulation and function". Reviews of Physiology, Biochemistry and Pharmacology 144: 1–94. 2002. doi:10.1007/BFb0116585. ISBN 978-3-540-42814-5. PMID 11987824.
- ↑ 3.0 3.1 "Kinetic disruption of lipid rafts is a mechanosensor for phospholipase D". Nature Communications 7 (1): 13873. December 2016. doi:10.1038/ncomms13873. PMID 27976674. Bibcode: 2016NatCo...713873P.
- ↑ 4.00 4.01 4.02 4.03 4.04 4.05 4.06 4.07 4.08 4.09 4.10 4.11 4.12 4.13 4.14 4.15 4.16 4.17 4.18 "Molecular structure of phospholipase D and regulatory mechanisms of its activity in plant and animal cells". Biochemistry. Biokhimiia 77 (1): 1–14. January 2012. doi:10.1134/S0006297912010014. PMID 22339628. https://nrc-publications.canada.ca/eng/view/accepted/?id=740b4fe5-17f1-4ef7-a4bf-ad2d609c57c2.
- ↑ 5.00 5.01 5.02 5.03 5.04 5.05 5.06 5.07 5.08 5.09 5.10 5.11 5.12 5.13 5.14 5.15 5.16 5.17 5.18 5.19 "Mammalian phospholipase D physiological and pathological roles". Acta Physiologica 204 (2): 219–26. February 2012. doi:10.1111/j.1748-1716.2011.02298.x. PMID 21447092.
- ↑ 6.0 6.1 6.2 6.3 6.4 6.5 6.6 6.7 6.8 "Phospholipase D in cell proliferation and cancer". Molecular Cancer Research 1 (11): 789–800. September 2003. PMID 14517341.
- ↑ "Phospholipase D Transduces Force to TREK-1 Channels in a Biological Membrane". bioRxiv: 758896. 5 September 2019. doi:10.1101/758896.
- ↑ "Studies on the mechanism of general anesthesia". Proceedings of the National Academy of Sciences of the United States of America 117 (24): 13757–13766. June 2020. doi:10.1073/pnas.2004259117. PMID 32467161. Bibcode: 2020PNAS..11713757P.
- ↑ 9.0 9.1 9.2 "Regulation and possible role of mammalian phospholipase D in cellular functions". Journal of Biochemistry 131 (3): 301–6. March 2002. doi:10.1093/oxfordjournals.jbchem.a003103. PMID 11872157.
- ↑ 10.0 10.1 10.2 10.3 "Phospholipase D". Biochemistry and Cell Biology 82 (1): 225–53. February 2004. doi:10.1139/o03-079. PMID 15052340.
- ↑ 11.0 11.1 "Protein kinase C alpha mediates phospholipase D activation by nucleotides and phorbol ester in Madin-Darby canine kidney cells. Stimulation of phospholipase D is independent of activation of polyphosphoinositide-specific phospholipase C and phospholipase A2". The Journal of Biological Chemistry 269 (14): 10511–6. April 1994. doi:10.1016/S0021-9258(17)34089-9. PMID 8144636.
- ↑ 12.0 12.1 12.2 "The first crystal structure of a phospholipase D". Structure 8 (6): 655–67. June 2000. doi:10.1016/S0969-2126(00)00150-7. PMID 10873862.
- ↑ "A role of p44/42 mitogen-activated protein kinases in formyl-peptide receptor-mediated phospholipase D activity and oxidant production". FASEB Journal 20 (1): 142–4. January 2006. doi:10.1096/fj.05-3881fje. PMID 16253958.
- ↑ "Phosphatidate accumulation in hormone-treated hepatocytes via a phospholipase D mechanism". The Journal of Biological Chemistry 262 (31): 15309–15. November 1987. doi:10.1016/S0021-9258(18)48176-8. PMID 3117799.
- ↑ "Ca2+-mobilizing hormones elicit phosphatidylethanol accumulation via phospholipase D activation". FEBS Letters 225 (1–2): 201–4. December 1987. doi:10.1016/0014-5793(87)81157-2. PMID 3319693.
- ↑ "Diacylglycerols and phosphatidates: which molecular species are intracellular messengers?". Trends in Biochemical Sciences 23 (6): 200–4. June 1998. doi:10.1016/S0968-0004(98)01200-6. PMID 9644971.
- ↑ "Cardiolipin synthase of Arabidopsis thaliana". FEBS Letters 579 (10): 2161–5. April 2005. doi:10.1016/j.febslet.2005.03.007. PMID 15811335.
- ↑ Nowicki M (2006). Characterization of the Cardiolipin Synthase from Arabidopsis thaliana (Ph.D. thesis). RWTH-Aachen University. Archived from the original on 2011-10-05. Retrieved 2011-07-11.
- ↑ "A novel family of phospholipase D homologues that includes phospholipid synthases and putative endonucleases: identification of duplicated repeats and potential active site residues". Protein Science 5 (5): 914–22. May 1996. doi:10.1002/pro.5560050513. PMID 8732763.
- ↑ "A duplicated catalytic motif in a new superfamily of phosphohydrolases and phospholipid synthases that includes poxvirus envelope proteins". Trends in Biochemical Sciences 21 (7): 242–3. July 1996. doi:10.1016/0968-0004(96)30024-8. PMID 8755242.
- ↑ "Cloning and expression of phosphatidylcholine-hydrolyzing phospholipase D from Ricinus communis L". The Journal of Biological Chemistry 269 (32): 20312–7. August 1994. doi:10.1016/S0021-9258(17)31993-2. PMID 8051126.
- ↑ "Regulation of eukaryotic phosphatidylinositol-specific phospholipase C and phospholipase D". Annual Review of Biochemistry 66: 475–509. 1997. doi:10.1146/annurev.biochem.66.1.475. PMID 9242915.
- ↑ 23.0 23.1 "Kinetic disruption of lipid rafts is a mechanosensor for phospholipase D". Nature Communications 7 (13873): 13873. December 2016. doi:10.1038/ncomms13873. PMID 27976674. Bibcode: 2016NatCo...713873P.
- ↑ "Studies on the mechanism of general anesthesia". bioRxiv 117 (24): 13757–13766. 4 May 2018. doi:10.1101/313973. PMID 32467161.
- ↑ 25.0 25.1 25.2 "Phospholipase D as a therapeutic target in brain disorders". Neuropsychopharmacology 37 (1): 301–2. January 2012. doi:10.1038/npp.2011.178. PMID 22157867.
- ↑ "Kinetic disruption of lipid rafts is a mechanosensor for phospholipase D". Nature Communications 7: 13873. December 2016. doi:10.1038/ncomms13873. PMID 27976674. Bibcode: 2016NatCo...713873P.
- ↑ 27.0 27.1 "A Molecular Target for an Alcohol Chain-Length Cutoff". Journal of Molecular Biology 431 (2): 196–209. January 2019. doi:10.1016/j.jmb.2018.11.028. PMID 30529033.
- ↑ "Synthesis of phosphatidylethanol--a potential marker for adult males at risk for alcoholism". Proceedings of the National Academy of Sciences of the United States of America 85 (24): 9778–82. December 1988. doi:10.1073/pnas.85.24.9778. PMID 3200856. Bibcode: 1988PNAS...85.9778M.
- ↑ "Lipid agonism: The PIP2 paradigm of ligand-gated ion channels". Biochimica et Biophysica Acta (BBA) - Molecular and Cell Biology of Lipids 1851 (5): 620–8. May 2015. doi:10.1016/j.bbalip.2015.01.011. PMID 25633344.
- ↑ "Phospholipase D2 specifically regulates TREK potassium channels via direct interaction and local production of phosphatidic acid". Proceedings of the National Academy of Sciences of the United States of America 111 (37): 13547–52. September 2014. doi:10.1073/pnas.1407160111. PMID 25197053. Bibcode: 2014PNAS..11113547C.
- ↑ "Rare coding variants in the phospholipase D3 gene confer risk for Alzheimer's disease". Nature 505 (7484): 550–554. January 2014. doi:10.1038/nature12825. PMID 24336208. Bibcode: 2014Natur.505..550..
External links
- Phospholipase+D at the US National Library of Medicine Medical Subject Headings (MeSH)
 |