Biology:Aldehyde deformylating oxygenase
| Aldehyde oxygenase (deformylating) | |||||||||
|---|---|---|---|---|---|---|---|---|---|
 | |||||||||
| Identifiers | |||||||||
| EC number | 4.1.99.5 | ||||||||
| Alt. names | Aldehyde decarbonylase | ||||||||
| Databases | |||||||||
| IntEnz | IntEnz view | ||||||||
| BRENDA | BRENDA entry | ||||||||
| ExPASy | NiceZyme view | ||||||||
| KEGG | KEGG entry | ||||||||
| MetaCyc | metabolic pathway | ||||||||
| PRIAM | profile | ||||||||
| PDB structures | RCSB PDB PDBe PDBsum | ||||||||
| |||||||||
Aldehyde deformylating oxygenases (ADO) (EC 4.1.99.5) are a family of enzymes which catalyze the oxygenation of medium and long chain aldehydes to alkanes via the removal of a carbonyl group as formate.
- n-aldehyde + O2 + 2 NADPH + H+ → (n-1)-alkane + formate + H2O + 2 NADP+
Aldehyde deformylating oxygenases are found in cyanobacteria as part of the alkane biosynthesis pathway.[2] Their substrates are medium- to long-chain aldehydes formed from acyl-ACP by acyl-ACP reductases (EC 1.2.1.80),[2] commonly of 16 and 18 carbons, but potentially as short as 9 carbons and 10 carbons.[3] Compared to other aldehyde decarbonylases, such as insect or plant aldehyde decarbonylase, cyanobacterial ADO is unusual in evolving formate rather than CO or CO2 and for residing in the cytosol.[3] It is also enzymatically unusual in catalyzing an formally hydrolytic and redox-neutral oxygenation of the substrate.[4]
Structure
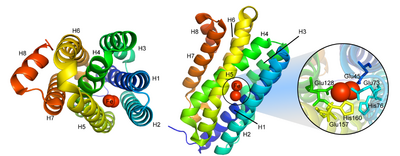
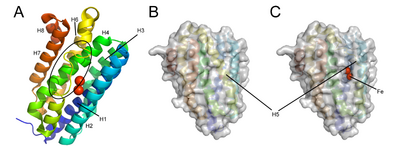
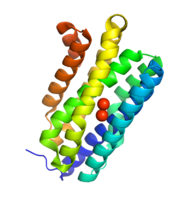
Cyanobacterial aldehyde deformylating oxygenases are cytosolic nonheme di-iron oxygenases, but are much smaller (29 kDa) than other members of the family,[3] and share sequence homology with ferritin-like or ribonucleotide reductases.[2] The overall structure is a bundle of 8 alpha-helices coordinating two central iron cofactors via histidine, aspartate and glutamate.[2] The substrate channels lies parallel to the helices and terminates at the di-iron center.[2]
Conformational changes during the enzymatic cycle of Synechococcus elongates ADO have been observed[5] (PDB: 4QUW, PDB: 4RC6, PDB: 4RC7, PDB: 4RC8). The binding of the substrate aldehyde displaces two coordinating residues on helix 5 (Glu157 and His160), causing a portion of the helix (residues 144-150) to unwind.[5] The resulting hole in the protein surface exposes the active site, facilitating the entrance of the cosubstrate oxygen.[5]
A similar conformational change has been observed for Prochlorococcus marinus ADO (PDB: 4PGI), in which residues 154-165 on helix 5 are unwound in the apoenzyme conformation to facilitate metal entry.[1]
Mechanism
The reaction catalyzed by ADO is unusual in that it is an oxygenation reaction which results in the formal hydrolysis, rather than oxidation, of the substrate.[4] The exact mechanism is not completely understood, and current understanding is based on a consensus between mechanistic studies and comparison with similar enzymes. The structurally similar R2 unit of ribonucleotide reductase proceeds via a tyrosyl radical mechanism, but the homologous tyrosine is replaced by phenylalanine in ADO.[2]
Mechanistic studies suggest that the aldehyde hydrogen is retained in the formate, the alkane hydrogen derives from the solvent, and one formate oxygen originates from O2.[6] The mechanism is tentatively hypothesized to take place by the following steps:[7]
- The reduced di-iron coordinates oxygen, which oxidizes the iron and forms a peroxide species.
- The peroxide species attacks the aldehyde.
- An electron transfer coupled with cleavage of the peroxo species generates a hemi-acetal radical.
- The terminal C-C bond cleaves homolytically to form the alkyl radical and release formate.
- The alkyl radical is quenched by final electron transfer.
- Two electron transfers restore the reduced state of di-iron and release a molecule of water.
Non-specific formation of alcohols rather than alkanes has also been observed, which would instead correspond to a heterolytic cleavage.[7]
Kinetics
The Km for O2 is 84 ± 9 µM.[8] However, the observed catalytic turnover is extremely inefficient, on the order of kcat = 1 min−1,[3] raising the possibility that the current understanding of the functional role, cofactors, or even substrates of ADO are incorrect. Transgenetically expressed, ADO appears to be dependent on ferredoxin-ferredoxin reductase to deliver reducing equivalents, but the endogenous reducing system is not known.[2] Further, oxygen-independent aldehyde deformylation has also been observed.[6]
H2O2 is an inhibitor of cADO, and an ADO-catalase fusion protein exhibits improved turnover.[8] Short-chain aldehydes are also observed to be substrate inhibitors.[6]
References
- ↑ 1.0 1.1 1.2 1.3 "Insights into Substrate and Metal Binding from the Crystal Structure of Cyanobacterial Aldehyde Deformylating Oxygenase with Substrate Bound". ACS Chem. Biol. 9 (11): 2584–2593. November 2014. doi:10.1021/cb500343j. PMID 25222710.
- ↑ 2.0 2.1 2.2 2.3 2.4 2.5 2.6 "Microbial biosynthesis of alkanes". Science 329 (5991): 559–562. Jul 2010. doi:10.1126/science.1187936. PMID 20671186. Bibcode: 2010Sci...329..559S.
- ↑ 3.0 3.1 3.2 3.3 "Aldehyde Decarbonylases: Enigmatic Enzymes of Hydrocarbon Biosynthesis". ACS Catal. 3 (11): 2515–2521. Sep 2013. doi:10.1021/cs400637t. PMID 24319622.
- ↑ 4.0 4.1 "Conversion of Fatty Aldehydes to Alka(e)nes and Formate by a Cyanobacterial Aldehyde Decarbonylase: Cryptic Redox by an Unusual Dimetal Oxygenase". J. Am. Chem. Soc. 133 (16): 6158–6161. Apr 2011. doi:10.1021/ja2013517. PMID 21462983.
- ↑ 5.0 5.1 5.2 "Structural insights into the catalytic mechanism of aldehyde-deformylating oxygenases". Protein & Cell 6 (1): 55–67. Jan 2015. doi:10.1007/s13238-014-0108-2. PMID 25482408.
- ↑ 6.0 6.1 6.2 "Oxygen-independent alkane formation by non-heme iron-dependent cyanobacterial aldehyde decarbonylase: investigation of kinetics and requirement for an external electron donor". Biochemistry 50 (49): 10743–50. Dec 2013. doi:10.1021/bi2012417. PMID 22074177.
- ↑ 7.0 7.1 "Rapid Reduction of the Diferric-Peroxyhemiacetal Intermediate in Aldehyde-Deformylating Oxygenase by a Cyanobacterial Ferredoxin: Evidence for a Free-Radical Mechanism". J. Am. Chem. Soc. 137 (36): 11695–11709. Aug 2015. doi:10.1021/jacs.5b06345. PMID 26284355.
- ↑ 8.0 8.1 "Fusing catalase to an alkane-producing enzyme maintains enzymatic activity by converting the inhibitory byproduct H2O2 to the cosubstrate O2". Proc. Natl. Acad. Sci. U.S.A. 110 (8): 3191–6. Feb 2013. doi:10.1073/pnas.1218769110. PMID 23391732. Bibcode: 2013PNAS..110.3191A.
 |