Biology:Beta-ketoacyl-ACP synthase III
| β-ketoacyl-(acyl-carrier-protein) synthase III | |||||||||
|---|---|---|---|---|---|---|---|---|---|
| Identifiers | |||||||||
| EC number | 2.3.1.180 | ||||||||
| CAS number | 9077-10-5 | ||||||||
| Databases | |||||||||
| IntEnz | IntEnz view | ||||||||
| BRENDA | BRENDA entry | ||||||||
| ExPASy | NiceZyme view | ||||||||
| KEGG | KEGG entry | ||||||||
| MetaCyc | metabolic pathway | ||||||||
| PRIAM | profile | ||||||||
| PDB structures | RCSB PDB PDBe PDBsum | ||||||||
| Gene Ontology | AmiGO / QuickGO | ||||||||
| |||||||||
| 3-Oxoacyl-[acyl-carrier-protein (ACP)] synthase III | |||||||||
|---|---|---|---|---|---|---|---|---|---|
 Structure and active-site architecture of beta-ketoacyl-acyl carrier protein synthase III (FabH) from escherichia coli.[1] | |||||||||
| Identifiers | |||||||||
| Symbol | ACP_syn_III | ||||||||
| Pfam | PF08545 | ||||||||
| InterPro | IPR013751 | ||||||||
| |||||||||
In enzymology, a β-ketoacyl-[acyl-carrier-protein] synthase III (EC 2.3.1.180) is an enzyme that catalyzes the chemical reaction
- acetyl-CoA + malonyl-[acyl carrier protein] [math]\displaystyle{ \rightleftharpoons }[/math] acetoacetyl-[acyl carrier protein] + CoA + CO2
Thus, the two substrates of this enzyme are acetyl-CoA and malonyl-[acyl-carrier-protein], whereas its 3 products are acetoacetyl-[acyl-carrier-protein], CoA, and CO2. This enzyme belongs to the family of transferases, to be specific those acyltransferases transferring groups other than aminoacyl groups.
This enzyme participates in fatty acid biosynthesis. β-Ketoacyl-acyl-carrier-protein synthase III is involved in the dissociated (or type II) fatty-acid biosynthesis system that occurs in plants and bacteria. The role of FabH in fatty acid synthesis has been described in Streptomyces glaucescens,[2] Streptococcus pneumoniae,[3] and Streptomyces coelicolor.[4]
Nomenclature
The systematic name of this enzyme class is acetyl-CoA:malonyl-[acyl-carrier-protein] C-acyltransferase. Other names in common use include:
|
|
Role in tuberculosis
Mycobacterium tuberculosis, the cause of tuberculosis, evades effective immune clearance through encapsulation, especially with mycolic acids that are particularly resistant to the normal degradative processes of macrophages. Furthermore, this capsule inhibits entry of antibiotics. The enzymes involved in mycolate biosynthesis are essential for survival and pathogenesis, and thus represent excellent drug targets.
In M. tuberculosis, the beta-ketoacyl-[acyl-carrier-protein] synthase III enzyme is designated mtFabH and is a crucial link between the fatty acid synthase-I and fatty acid synthase-II pathways producing mycolic acids. FAS-I is involved in the synthesis of C16 and C26 fatty acids. The C16 acyl-CoA product acts as a substrate for the synthesis of meromycolic acid by FAS-II, whereas the C26 fatty acid constitutes the alpha branch of the final mycolic acid. MtFabH has been proposed to be the link between FAS-I and FAS-II by converting C14-CoA generated by FAS-I to C16-AcpM, which is channelled into the FAS-II cycle.[5] According to in silico flux balance analyses,[6] mtFabH is essential but not according to transposon site hybridization analysis.[7] Unlike the enzymes in FAS-I, the enzymes of FAS-II, including mtFabH, and are not found in mammals, suggesting inhibitors of these enzymes are suitable choices for drug development.
Structure and substrates
Crystal structures of FabH have been reported from Mycobacterium tuberculosis,[1][8][9] Staphylococcus aureus,[10] Escherichia coli,[11] and Thermus thermophilus.[12]
The catalytic activity and substrate specificity of mtFabH has been measured[13] then further probed using crystallographic and directed mutagenesis methods.[14] Structures have been determined of ecFabH bound with substrates, (CoA, malonyl CoA, degraded CoA).[8] Specific inhibitors developed using rational design have recently been reported.[15][16][17] In 2005, the structure of a catalytically disabled mtFabH mutant with lauroyl-CoA was reported.[18]
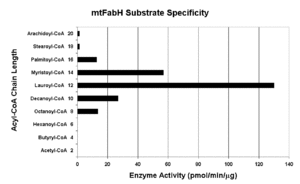
Native mtFabH is a homodimer with Mr = 77 ± 25 kDa. Although there is substantial structural homology among all bacterial FabH enzymes determined thus far, with two channels for binding of acyl-CoA and malonyl-ACP substrates and a conserved catalytic triad (C122, H258, N289 in mtFabH), mtFabH contains residues along the acyl-CoA binding channel that preferentially select for longer-chain substrates peaking with lauroyl-CoA (C12). Inhibition strategies based on rational design could include competitive displacement of the substrates or disruption of the catalytic site. Phosphorylation of Thr45, which is located at the entrance of the substrate channel, inhibits activity, perhaps by altering accessibility of substrates.[19]
Inhibitors
At least two of the existing drugs for tuberculosis were originally derived from microbes; cerulenin from the fungus Cephalosporium caerulens and thiolactomycin (TLM) from the actinomycete Nocardia spp. Isoniazid (isonicotinic acid hydrazide), ethionamide, triclosan [5-chloro-2-(2,4-dichlorophenoxy)-phenol] and TLM are known to specifically inhibit mycolic acid biosynthesis.[20] Derivatives of TLM and related compounds are being screened to improve efficacy.[21][22][23][24]
While much has been learned from these structural studies and rational design is an excellent approach to develop novel inhibitors, alternative approaches such as bio-prospecting may reveal unexpected compounds such as an allosteric inhibitor discovered by Daines et al. This could be especially important given that phosphorylation of mycolate synthesis enzymes is suggested to be critical to regulation and kinase domains are known to have multiple control mechanisms remote from ligand binding and active sites.[19]
Following the discovery that phomallenic acids isolated from a leaf litter fungus identified as Phoma sp. are inhibitors of the FabH/FabF.[25][26] Wang et al. recently reported their discovery from the soil bacterium Streptomyces platensis of a novel natural inhibitor of FabH with in vivo activity called platencin.[27] These were found by screening 250,000 extracts of soil bacteria and fungi, demonstrating the viability of bio-prospecting. While a potentially useful antibiotic in its own right, it has now been shown that platensimycin is not specifically active on mtFabH.[28]
It is speculated that novel inhibitors will most likely be small molecules of relatively low polarity, considering that the catalytic sites of the mtFabH homodimer are hidden in relatively hydrophobic pockets and the need to traverse capsules of established bacilli. This is supported by the poor water solubility of an inhibitor to ecFabH. It is also hoped that, by being small molecules, their synthesis or biosynthesis will be simple and cheap, thereby enhancing affordability of subsequent drugs to developing countries. Techniques for screening efficacy of inhibitors are available.[29][30]
Therapeutic potential
In 2005, tuberculosis caused approximately 1.6 million deaths worldwide, 8.8 million people became sick, with 90% of these cases in developing countries, and an estimated one-third of the world's population has latent TB.[31][32] Despite the availability of the BCG vaccine and multiple antibiotics, until 2005 TB resurged due to multidrug resistance, exacerbated by incubation in immune-compromised AIDS victims, drug treatment non-compliance, and ongoing systemic deficiencies of healthcare in developing countries. Mortality and infection rates appear to have peaked, but TB remains a serious global problem. New effective drugs are needed to combat this disease. Inhibitors against mtFabH, or against other enzymes of the FAS-II pathway, may have broader utility, such as the treatment of multidrug-resistant Staphylococcus aureus, and Plasmodium falciparum, the causative agent of another serious refractory problem, malaria.
Given the predominance of TB in poor countries, the commercial incentive to develop new drugs has been hampered, along with complacency and reliance on old, well-established, "first-line" drugs such as Rifampicin, Isoniazid, Pyrazinamide, and Ethambutol. The price point is already very low: US$16–35 will buy a full six-month drug course[33] Nevertheless, new drugs are in clinical trials.[34][35]
According to the Global Alliance for TB Drug Development, sales of first-line TB drugs are projected to be approximately US$315 million per year, and US$54 million for second-line treatments, yet the global economic toll of TB is at least $12 billion each year.[36][37]
References
- ↑ Jump up to: 1.0 1.1 "The 1.8 A crystal structure and active-site architecture of beta-ketoacyl-acyl carrier protein synthase III (FabH) from escherichia coli". Structure 8 (2): 185–95. 2000. doi:10.1016/S0969-2126(00)00094-0. PMID 10673437.
- ↑ "Characterization of β-Ketoacyl-Acyl Carrier Protein Synthase III from Streptomyces glaucescens and Its Role in Initiation of Fatty Acid Biosynthesis". J. Bacteriol. 180 (17): 4481–6. 1998. doi:10.1128/JB.180.17.4481-4486.1998. PMID 9721286.
- ↑ "Identification, substrate specificity, and inhibition of the Streptococcus pneumoniae beta-ketoacyl-acyl carrier protein synthase III (FabH)". J. Biol. Chem. 276 (32): 30024–30. 2001. doi:10.1074/jbc.M101769200. PMID 11375394.
- ↑ "Alteration of the Fatty Acid Profile of Streptomyces coelicolor by Replacement of the Initiation Enzyme 3-Ketoacyl Acyl Carrier Protein Synthase III (FabH)". J. Bacteriol. 187 (11): 3795–9. 2005. doi:10.1128/JB.187.11.3795-3799.2005. PMID 15901703.
- ↑ "The Mycobacterium tuberculosis FAS-II condensing enzymes: their role in mycolic acid biosynthesis, acid-fastness, pathogenesis and in future drug development". Mol. Microbiol. 64 (6): 1442–54. June 2007. doi:10.1111/j.1365-2958.2007.05761.x. PMID 17555433.
- ↑ "Flux Balance Analysis of Mycolic Acid Pathway: Targets for Anti-Tubercular Drugs". PLOS Comput. Biol. 1 (5): e46. October 2005. doi:10.1371/journal.pcbi.0010046. PMID 16261191. Bibcode: 2005PLSCB...1...46R.
- ↑ "Genes required for mycobacterial growth defined by high density mutagenesis". Mol. Microbiol. 48 (1): 77–84. April 2003. doi:10.1046/j.1365-2958.2003.03425.x. PMID 12657046.
- ↑ Jump up to: 8.0 8.1 PDB: 1HND, 1HNH, 1HNJ; "Refined structures of beta-ketoacyl-acyl carrier protein synthase III". J. Mol. Biol. 307 (1): 341–56. March 2001. doi:10.1006/jmbi.2000.4457. PMID 11243824.
- ↑ PDB: 1HZP; "Crystal structure of the Mycobacterium tuberculosis beta-ketoacyl-acyl carrier protein synthase III". J. Biol. Chem. 276 (23): 20516–22. June 2001. doi:10.1074/jbc.M010762200. PMID 11278743.
- ↑ PDB: 1ZOW; "Crystal structure and substrate specificity of the β-ketoacyl-acyl carrier protein synthase III (FabH) from Staphylococcus aureus". Protein Sci. 14 (8): 2087–94. August 2005. doi:10.1110/ps.051501605. PMID 15987898.
- ↑ PDB: 1HN9; "Crystal structure of beta-ketoacyl-acyl carrier protein synthase III. A key condensing enzyme in bacterial fatty acid biosynthesis". J. Biol. Chem. 274 (51): 36465–71. December 1999. doi:10.1074/jbc.274.51.36465. PMID 10593943.
- ↑ PDB: 1UB7Inagaki E, Kuramitsu S, Yokoyama S, Miyano M, Tahirov TH (2007) The Crystal Structure of Beta-Ketoacyl-[Acyl Carrier Protein] Synthase III (Fabh) from Thermus thermophilus.
- ↑ "Identification and substrate specificity of beta -ketoacyl (acyl carrier protein) synthase III (mtFabH) from Mycobacterium tuberculosis". J. Biol. Chem. 275 (36): 28201–7. September 2000. doi:10.1074/jbc.M003241200. PMID 10840036.
- ↑ PDB: 1M1M, 2AJ9; "Probing the mechanism of the Mycobacterium tuberculosis beta-ketoacyl-acyl carrier protein synthase III mtFabH: factors influencing catalysis and substrate specificity". J. Biol. Chem. 280 (37): 32539–47. September 2005. doi:10.1074/jbc.M413216200. PMID 16040614.
- ↑ PDB: 1MZS; "First X-ray cocrystal structure of a bacterial FabH condensing enzyme and a small molecule inhibitor achieved using rational design and homology modeling". J. Med. Chem. 46 (1): 5–8. January 2003. doi:10.1021/jm025571b. PMID 12502353.
- ↑ "Structure-based design, synthesis, and study of potent inhibitors of beta-ketoacyl-acyl carrier protein synthase III as potential antimicrobial agents". J. Med. Chem. 48 (5): 1596–609. March 2005. doi:10.1021/jm049141s. PMID 15743201.
- ↑ "A combined approach of docking and 3D QSAR study of beta-ketoacyl-acyl carrier protein synthase III (FabH) inhibitors". Bioorg. Med. Chem. 14 (5): 1474–82. March 2006. doi:10.1016/j.bmc.2005.10.001. PMID 16275103.
- ↑ PDB: 1U6S; "Crystal structure of a substrate complex of Mycobacterium tuberculosis beta-ketoacyl-acyl carrier protein synthase III (FabH) with lauroyl-coenzyme A". J. Mol. Biol. 346 (5): 1313–21. March 2005. doi:10.1016/j.jmb.2004.12.044. PMID 15713483.
- ↑ Jump up to: 19.0 19.1 "The Mycobacterium tuberculosis β-Ketoacyl-Acyl Carrier Protein Synthase III Activity Is Inhibited by Phosphorylation on a Single Threonine Residue". J. Biol. Chem. 284 (10): 6414–24. March 2009. doi:10.1074/jbc.M806537200. PMID 19074144.
- ↑ "Drugs that inhibit mycolic acid biosynthesis in Mycobacterium tuberculosis". Curr Pharm Biotechnol 3 (3): 197–225. September 2002. doi:10.2174/1389201023378328. PMID 12164478.
- ↑ "Biphenyl-based analogues of thiolactomycin, active against Mycobacterium tuberculosis mtFabH fatty acid condensing enzyme". Bioorg. Med. Chem. Lett. 13 (21): 3685–8. November 2003. doi:10.1016/j.bmcl.2003.08.015. PMID 14552758.
- ↑ "Acetylene-based analogues of thiolactomycin, active against Mycobacterium tuberculosis mtFabH fatty acid condensing enzyme". Bioorg. Med. Chem. Lett. 14 (2): 373–6. January 2004. doi:10.1016/j.bmcl.2003.10.061. PMID 14698162.
- ↑ "1,2-Dithiole-3-Ones as Potent Inhibitors of the Bacterial 3-Ketoacyl Acyl Carrier Protein Synthase III (FabH)". Antimicrob. Agents Chemother. 48 (8): 3093–102. August 2004. doi:10.1128/AAC.48.8.3093-3102.2004. PMID 15273125.
- ↑ Todd, Matthew H., ed (2009). "Identification of 2-Aminothiazole-4-Carboxylate Derivatives Active against Mycobacterium tuberculosis H37Rv and the β-Ketoacyl-ACP Synthase mtFabH". PLOS ONE 4 (5): e5617. doi:10.1371/journal.pone.0005617. PMID 19440303. Bibcode: 2009PLoSO...4.5617A.
- ↑ "Discovery of FabH/FabF Inhibitors from Natural Products". Antimicrob. Agents Chemother. 50 (2): 519–26. February 2006. doi:10.1128/AAC.50.2.519-526.2006. PMID 16436705.
- ↑ "Discovery of bacterial fatty acid synthase inhibitors from a Phoma species as antimicrobial agents using a new antisense-based strategy". J. Nat. Prod. 69 (3): 377–80. March 2006. doi:10.1021/np050416w. PMID 16562839.
- ↑ "Discovery of platencin, a dual FabF and FabH inhibitor with in vivo antibiotic properties". Proceedings of the National Academy of Sciences of the United States of America 104 (18): 7612–6. May 2007. doi:10.1073/pnas.0700746104. PMID 17456595. Bibcode: 2007PNAS..104.7612W.
- ↑ Ahmed, Niyaz, ed (2009). "Platensimycin Activity against Mycobacterial β-Ketoacyl-ACP Synthases". PLOS ONE 4 (7): e6306. doi:10.1371/journal.pone.0006306. PMID 19609444. Bibcode: 2009PLoSO...4.6306B.
- ↑ "Mycolic Acid Index Susceptibility Method for Mycobacterium tuberculosis". J. Clin. Microbiol. 39 (7): 2642–5. July 2001. doi:10.1128/JCM.39.7.2642-2645.2001. PMID 11427584.
- ↑ "Development of a scintillation proximity assay for beta-ketoacyl-acyl carrier protein synthase III". Anal. Biochem. 282 (1): 107–14. June 2000. doi:10.1006/abio.2000.4594. PMID 10860506.
- ↑ "The growing burden of tuberculosis: global trends and interactions with the HIV epidemic". Arch. Intern. Med. 163 (9): 1009–21. May 2003. doi:10.1001/archinte.163.9.1009. PMID 12742798. https://researchonline.lshtm.ac.uk/id/eprint/17507/1/The%20Growing%20Burden%20of%20Tuberculosis.pdf.
- ↑ "Global Tuberculosis Control 2007". World Health Organization. 2007. Archived from the original on 2010-02-02. https://web.archive.org/web/20100202185311/http://www.who.int/tb/publications/global_report/2007/download_centre/en/index.html. Retrieved 2010-01-02.
- ↑ "TB Alliance — An Outdated Treatment". Global Alliance for TB Drug Development. Archived from the original on 2010-01-13. https://web.archive.org/web/20100113165524/http://www.tballiance.org/why/outdated.php. Retrieved 2010-01-02.
- ↑ "New Approaches to Filling the Gap in Tuberculosis Drug Discovery". PLOS Med. 4 (11): e293. November 2007. doi:10.1371/journal.pmed.0040293. PMID 17988169.
- ↑ "TB Alliance — TB Drug Portfolio". Global Alliance for TB Drug Development. Archived from the original on 2010-01-13. https://web.archive.org/web/20100113155341/http://www.tballiance.org/new/portfolio.php. Retrieved 2010-01-02.
- ↑ "New Study Reveals Limitations of a Complex and Challenging Global Tuberculosis Drug Marketplace". TB Alliance Newscenter: News Release. Global Alliance for TB Drug Development. 2007-05-14. http://www.tballiance.org/newscenter/view-brief.php?id=683. Retrieved 2010-01-02.
- ↑ "The Economics of TB Drug Development". Global Alliance for TB Drug Development. 2001. http://www.tballiance.org/downloads/publications/TBA_Economics_Report.pdf. Retrieved 2010-01-02.
Further reading
- "Isolation and characterization of the beta-ketoacyl-acyl carrier protein synthase III gene (fabH) from Escherichia coli K-12". J. Biol. Chem. 267 (10): 6807–14. 1992. doi:10.1016/S0021-9258(19)50498-7. PMID 1551888.
- Overview of all the structural information available in the PDB for UniProt: P9WNG3 (Mycobacterium tuberculosis 3-oxoacyl-[acyl-carrier-protein] synthase 3) at the PDBe-KB.
 |