Biology:Carbohydrate sulfotransferase
| Carbohydrate Sulfotransferase family 2 | |||||||||
|---|---|---|---|---|---|---|---|---|---|
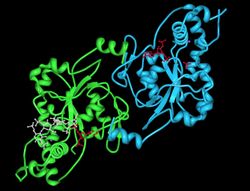 Example carbohydrate sulfotransferase with PAPS cosubstrate and carbohydrate substrate: Crystal Structure of human 3-O-Sulfotransferase-3 with bound PAPS and tetrasaccharide substrate. Enzyme chain A (blue), Enzyme chain B (green), PAPS (red), tetrasaccharide substrate (white), sodium ion (purple sphere).[1] | |||||||||
| Identifiers | |||||||||
| Symbol | Sulfotransfer_2 | ||||||||
| Pfam | PF03567 | ||||||||
| InterPro | IPR005331 | ||||||||
| Membranome | 495 | ||||||||
| |||||||||
| Carbohydrate Sulfotransferase family 1 | |
|---|---|
| Identifiers | |
| Symbol | Sulfotransfer_1 |
| InterPro | IPR016469 |
| Membranome | 493 |
In biochemistry, carbohydrate sulfotransferases are enzymes within the class of sulfotransferases which catalyze the transfer of the sulfate (–SO−
3) functional group to carbohydrate groups in glycoproteins and glycolipids. Carbohydrates are used by cells for a wide range of functions from structural purposes to extracellular communication. Carbohydrates are suitable for such a wide variety of functions due to the diversity in structure generated from monosaccharide composition, glycosidic linkage positions, chain branching, and covalent modification.[2] Possible covalent modifications include acetylation, methylation, phosphorylation, and sulfation.[3] Sulfation, performed by carbohydrate sulfotransferases, generates carbohydrate sulfate esters (–O–SO−
3). These sulfate esters are only located extracellularly, whether through excretion into the extracellular matrix (ECM) or by presentation on the cell surface.[4] As extracellular compounds, sulfated carbohydrates are mediators of intercellular communication, cellular adhesion, and ECM maintenance.
Enzyme mechanism
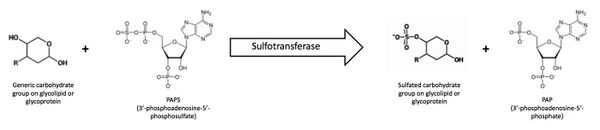
Sulfotransferases catalyze the transfer of a sulfonyl group from an activated sulfate donor onto a hydroxyl group (or an amino group, although this is less common) of an acceptor molecule.[4] In eukaryotic cells the activated sulfate donor is 3'-phosphoadenosine-5'-phosphosulfate (PAPS) (Figure 1).[5]
PAPS is synthesized in the cytosol from ATP and sulfate through the sequential action of ATP sulfurylase and APS kinase.[6] ATP sulfurylase first generates adenosine-5'-phosphosulfate (APS) and then APS kinase transfers a phosphate from ATP to APS to create PAPS. The importance of PAPS and sulfation has been discerned in previous studies by using chlorate, an analogue of sulfate, as a competitive inhibitor of ATP sulfurylase.[7] PAPS is a cosubstrate and source of activated sulfate for both cytosolic sulfotransferases and carbohydrate sulfotransferases, which are located in the Golgi. PAPS moves between the cytosol and the Golgi lumen via PAPS/PAP (3’-phosphoadenosine-5’-phosphate) translocase, a transmembrane antiporter.[8]
The exact mechanism used by sulfotransferases is still being elucidated, but studies have indicated that sulfotransferases use an in-line sulfonyl-transfer mechanism that is analogous to the phosphoryl transfer mechanism used by many kinases, which is logical given the great level of structural and functional similarities between kinases and sulfotransferases (Figure 2).[9] In carbohydrate sulfotransferases a conserved lysine has been identified in the active PAPS binding site, which is analogous to a conserved lysine in the active ATP binding site of kinases.[10][11] Protein sequence alignment studies indicate that this lysine is conserved in cytosolic sulfotransferases as well.[4]
In addition to the conserved lysine, sulfotransferases have a highly conserved histidine in the active site.[12] Based on the conservation of these residues, theoretical models, and experimental measurements a theoretical transition state for catalyzed sulfation has been proposed (Figure 3).[12]
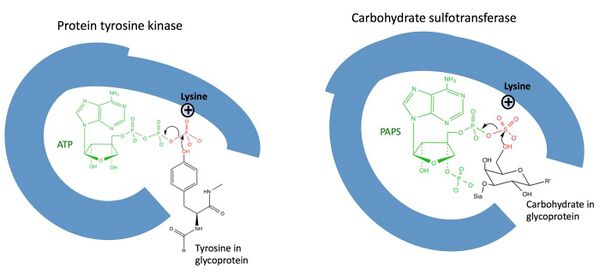 Figure 2: The mechanism by which carbohydrate sulfotransferase catalyzes the transfer of a sulfonyl group to a carbohydrate group in a glycoprotein or glycolipid is analogous to the mechanism by which a kinase catalyzes a phosphoryl group. Both enzymes use a lysine residue in their active sites to coordinate to their cosubstrates; the ATP cosubstrate in the kinase mechanism is analogous to the PAPS in the carbohydrate sulfotransferase mechanism (green). Red shows the group being transferred; note that the transfer is coordinated around the lysine. Black is the substrate. Sia stands for sialic acid.[6][9] |
 Figure 3: Transition state for catalyzed sulfation as proposed by Chapman et al. 2004.[12] Note the use of the conserved lysine and histidine residues. |
Biological function
Carbohydrate sulfotransferases are transmembrane enzymes in the Golgi that modify carbohydrates on glycolipids or glycoproteins as they move along the secretory pathway.[4] They have a short cytoplasmic N-terminal, one transmembrane domain, and a large C-terminal Golgi luminal domain.[6] They are distinct from cytosolic sulfotransferases in both structure and function. While cytosolic sulfotransferases play a metabolic role by modifying small molecule substrates such as steroids, flavonoids, neurotransmitters, and phenols, carbohydrate sulfotransferases have a fundamental role in extracellular signalling and adhesion by generating unique ligands through the modification of carbohydrate scaffolds.[4][13] Since the substrates of carbohydrate sulfotransferases are larger, they have larger active sites than cytosolic sulfotransferases.
There are two major families of carbohydrate sulfotransferases: heparan sulfotransferases and galactose/N-acetylgalactosamine/N-acetylglucosamine 6-O-sulfotransferases (GSTs).[14][15]
Heparan Sulfotransferases
Heparan sulfate is a glycosaminoglycan (GAG) that is linked by xylose to serine residues of proteins such as perlecan, syndecan, or glypican.[16] Sulfation of heparan sulfate GAGs helps give diversity to cell surface proteins and provides them with a unique sulfation pattern that allows them to specifically interact with other proteins.[12] For example, in mast cells the AT-III-binding pentasaccharide is synthesized with essential heparan sulfate sulfation steps. The binding of the heparan sulfate in this pentasaccharide to AT-III inactivates the blood-coagulation factors thrombin and Factor Xa.[17] Heparan sulfates are also known to interact with growth factors, cytokines, chemokines, lipid and membrane binding proteins, and adhesion molecules.[12]
GSTs
GSTs catalyze sulfation at the 6-hydroxyl group of galactose, N-acetylgalactosamine, or N-acetylglucosamine.[15] Like heparan sulfotransferases, GSTs are responsible for post-translational protein sulfation that assists in cell-signaling. GSTs are also responsible for the sulfation of extracellular matrix (ECM) proteins that assist with maintaining the structure between cells[4][18] For example, GSTs catalyze the sulfation of glycoproteins displaying the L-selectin binding epitope 6-sulfo sialyl Lewis x, which recruits leukocytes to areas of chronic inflammation.[18] GSTs are also responsible for the proper function of the ECM in the cornea; improper sulfation by GSTs can lead to opaque corneas.[18]
Disease Relevance
Carbohydrate sulfotransferases are of great interest as drug targets because of their essential roles in cell-cell signalling, adhesion, and ECM maintenance. Their roles in blood coagulation, chronic inflammation, and cornea maintenance mentioned in the Biological Function section above are all of interest for potential therapeutic purposes. In addition to these roles, carbohydrate sulfotransferases are of pharmacological interest because of their roles in viral infection, including herpes simplex virus 1 (HSV-1) and human immunodeficiency virus 1 (HIV-1).[12] Heparan sulfate sites have been shown to be essential for HSV-1 binding that leads to the virus entering the cell.[19] In contrast, heparan sulfate complexes have been shown to bind to HIV-1 and prevent it from entering the cell through its intended target, the CD4 receptor.[12]
Mutation in Carbohydrate sulfotransferases 6 (CHST6) is associated with Macular Corneal Dystrophy (MCD) Inheritance: Autosomal recessive. Genetic Locus: 16q22 Online Mendelian Inheritance in man (OMIM) Entry OMIM #217800
Human proteins from this family
- Carbohydrate sulfotransferases 6 (CHST6) Sulfotransferase that utilizes 3'-phospho-5'-adenylyl sulfate (PAPS) as sulfonate donor to catalyze the transfer of sulfate to position 6 of non-reducing N-acetylglucosamine (GlcNAc) residues of keratan. Mediates sulfation of keratan in cornea. Keratan sulfate plays a central role in maintaining corneal transparency.
- Carbohydrate sulfotransferases 8 (CHST8) and 9 (CHST9), which transfer sulfate to position 4 of non-reducing N-acetylgalactosamine (GalNAc) residues in both N-glycans and O-glycans.[20] They function in the biosynthesis of glycoprotein hormones lutropin and thyrotropin, by mediating sulfation of their carbohydrate structures.
- Carbohydrate sulfotransferase 10 (CHST10), which transfers sulfate to position 3 of the terminal glucuronic acid in both protein- and lipid-linked oligosaccharides.[21] It directs the biosynthesis of the HNK-1 carbohydrate structure, a sulfated glucuronyl-lactosaminyl residue carried by many neural recognition molecules, which is involved in cell interactions during ontogenetic development and in synaptic plasticity in the adult.
- Carbohydrate sulfotransferases 11 - 13 (CHST11, CHST12, CHST13), which catalyze the transfer of sulfate to position 4 of the GalNAc residue of chondroitin.[22] Chondroitin sulfate constitutes the predominant proteoglycan present in cartilage and is distributed on the surfaces of many cells and extracellular matrices. Some, thought not all, of these enzymes also transfer sulfate to dermatan.
- Carbohydrate sulfotransferase D4ST1 (D4ST1), which transfers sulfate to position 4 of the GalNAc residue of dermatan sulfate.[23]
References
- ↑ Moon, AF.; Edavettal, SC.; Krahn, JM.; Munoz, EM.; Negishi, M.; Linhardt, RJ.; Liu, J.; Pedersen, LC. (Oct 2004). "Structural analysis of the sulfotransferase (3-o-sulfotransferase isoform 3) involved in the biosynthesis of an entry receptor for herpes simplex virus 1". J Biol Chem 279 (43): 45185–93. doi:10.1074/jbc.M405013200. PMID 15304505.
- ↑ Nelson, R M; Venot, A; Bevilacqua, M P; Linhardt, R J; Stamenkovic, I (1995). "Carbohydrate-Protein Interactions in Vascular Biology". Annual Review of Cell and Developmental Biology 11 (1): 601–631. doi:10.1146/annurev.cb.11.110195.003125. ISSN 1081-0706. PMID 8689570.
- ↑ Hooper, L.V.; Baenziger, J.U. (1993). "Sulfotransferase and Glycosyltransferase Analyses Using a 96-Well Filtration Plate". Analytical Biochemistry 212 (1): 128–133. doi:10.1006/abio.1993.1301. ISSN 0003-2697. PMID 8368484.
- ↑ 4.0 4.1 4.2 4.3 4.4 4.5 Bowman, K; Bertozzi, C (1999). "Carbohydrate sulfotransferases: mediators of extracellular communication". Chemistry & Biology 6 (1): R9–R22. doi:10.1016/S1074-5521(99)80014-3. ISSN 1074-5521. PMID 9889154.
- ↑ Klaassen, CD.; Boles, JW. (May 1997). "Sulfation and sulfotransferases 5: the importance of 3'-phosphoadenosine 5'-phosphosulfate (PAPS) in the regulation of sulfation". FASEB J 11 (6): 404–18. doi:10.1096/fasebj.11.6.9194521. PMID 9194521.
- ↑ 6.0 6.1 6.2 Hemmerich, Stefan (2001). "Carbohydrate sulfotransferases: novel therapeutic targets for inflammation, viral infection and cancer". Drug Discovery Today 6 (1): 27–35. doi:10.1016/S1359-6446(00)01581-6. ISSN 1359-6446. PMID 11165170.
- ↑ Baeuerle, PA.; Huttner, WB. (Dec 1986). "Chlorate--a potent inhibitor of protein sulfation in intact cells". Biochem Biophys Res Commun 141 (2): 870–7. doi:10.1016/s0006-291x(86)80253-4. PMID 3026396.
- ↑ Ozeran, JD.; Westley, J.; Schwartz, NB. (Mar 1996). "Identification and partial purification of PAPS translocase". Biochemistry 35 (12): 3695–703. doi:10.1021/bi951303m. PMID 8619989.
- ↑ 9.0 9.1 Kakuta, Y.; Petrotchenko, EV.; Pedersen, LC.; Negishi, M. (Oct 1998). "The sulfuryl transfer mechanism. Crystal structure of a vanadate complex of estrogen sulfotransferase and mutational analysis". J Biol Chem 273 (42): 27325–30. doi:10.1074/jbc.273.42.27325. PMID 9765259.
- ↑ Kamio, K.; Honke, K.; Makita, A. (Dec 1995). "Pyridoxal 5'-phosphate binds to a lysine residue in the adenosine 3'-phosphate 5'-phosphosulfate recognition site of glycolipid sulfotransferase from human renal cancer cells". Glycoconj J 12 (6): 762–6. doi:10.1007/bf00731236. PMID 8748152.
- ↑ Sueyoshi, Tatsuya; Kakuta, Yoshimitsu; Pedersen, Lars C.; Wall, Frances E.; Pedersen, Lee G.; Negishi, Masahiko (1998). "A role of Lys614 in the sulfotransferase activity of human heparan sulfate N-deacetylase/N-sulfotransferase". FEBS Letters 433 (3): 211–214. doi:10.1016/S0014-5793(98)00913-2. ISSN 0014-5793. PMID 9744796.
- ↑ 12.0 12.1 12.2 12.3 12.4 12.5 12.6 Chapman, Eli; Best, Michael D.; Hanson, Sarah R.; Wong, Chi-Huey (2004). "Sulfotransferases: Structure, Mechanism, Biological Activity, Inhibition, and Synthetic Utility". Angewandte Chemie International Edition 43 (27): 3526–3548. doi:10.1002/anie.200300631. ISSN 1433-7851. PMID 15293241.
- ↑ Falany, CN. (Mar 1997). "Enzymology of human cytosolic sulfotransferases". FASEB J 11 (4): 206–16. doi:10.1096/fasebj.11.4.9068609. PMID 9068609.
- ↑ Shworak, NW.; Liu, J.; Petros, LM.; Zhang, L.; Kobayashi, M.; Copeland, NG.; Jenkins, NA.; Rosenberg, RD. (Feb 1999). "Multiple isoforms of heparan sulfate D-glucosaminyl 3-O-sulfotransferase. Isolation, characterization, and expression of human cdnas and identification of distinct genomic loci". J Biol Chem 274 (8): 5170–84. doi:10.1074/jbc.274.8.5170. PMID 9988767.
- ↑ 15.0 15.1 Hemmerich, S.; Rosen, SD. (Sep 2000). "Carbohydrate sulfotransferases in lymphocyte homing". Glycobiology 10 (9): 849–56. doi:10.1093/glycob/10.9.849. PMID 10988246.
- ↑ Rosenberg, RD.; Shworak, NW.; Liu, J.; Schwartz, JJ.; Zhang, L. (May 1997). "Heparan sulfate proteoglycans of the cardiovascular system. Specific structures emerge but how is synthesis regulated?". J Clin Invest 99 (9): 2062–70. doi:10.1172/JCI119377. PMID 9151776.
- ↑ Liu, J.; Shworak, NW.; Fritze, LM.; Edelberg, JM.; Rosenberg, RD. (Oct 1996). "Purification of heparan sulfate D-glucosaminyl 3-O-sulfotransferase". J Biol Chem 271 (43): 27072–82. doi:10.1074/jbc.271.43.27072. PMID 8900198.
- ↑ 18.0 18.1 18.2 Grunwell, Jocelyn R.; Rath, Virginia L.; Rasmussen, Jytte; Cabrilo, Zeljka; Bertozzi, Carolyn R. (2002). "Characterization and Mutagenesis of Gal/GlcNAc-6-O-sulfotransferases†". Biochemistry 41 (52): 15590–15600. doi:10.1021/bi0269557. ISSN 0006-2960. PMID 12501187.
- ↑ Shukla, D.; Liu, J.; Blaiklock, P.; Shworak, NW.; Bai, X.; Esko, JD.; Cohen, GH.; Eisenberg, RJ. et al. (Oct 1999). "A novel role for 3-O-sulfated heparan sulfate in herpes simplex virus 1 entry". Cell 99 (1): 13–22. doi:10.1016/S0092-8674(00)80058-6. PMID 10520990.
- ↑ "R in both N- and O-glycans". Glycobiology 11 (6): 495–504. 2001. doi:10.1093/glycob/11.6.495. PMID 11445554.
- ↑ "Expression cloning of a human sulfotransferase that directs the synthesis of the HNK-1 glycan on the neural cell adhesion molecule and glycolipids". J. Biol. Chem. 273 (9): 5190–5. 1998. doi:10.1074/jbc.273.9.5190. PMID 9478973.
- ↑ "Molecular cloning and expression of two distinct human chondroitin 4-O-sulfotransferases that belong to the HNK-1 sulfotransferase gene family". J. Biol. Chem. 275 (26): 20188–96. 2000. doi:10.1074/jbc.M002443200. PMID 10781601.
- ↑ "Molecular cloning and characterization of a dermatan-specific N-acetylgalactosamine 4-O-sulfotransferase". J. Biol. Chem. 276 (39): 36344–53. 2001. doi:10.1074/jbc.M105848200. PMID 11470797.
External links
 |