Biology:Polyamine oxidase
A polyamine oxidase (PAO) is an enzymatic flavoprotein that oxidizes a carbon-nitrogen bond in a secondary amino group of a polyamine donor, using molecular oxygen as an acceptor. The generalized PAO reaction converts three substrates (water, oxygen, and a polyamine with both primary and secondary amino groups) into three products (hydrogen peroxide, an amino-aldehyde, and a primary amine). Different PAOs with varying substrate specificities exist in different organisms. Phylogenetic analyses suggest that PAOs likely evolved once in eukaryotes and diversified by divergent evolution and gene duplication events, though some prokaryotes have acquired PAOs through horizontal gene transfer.[1]
| Substrate | Amino-aldehyde Product | Primary Amine Product | EC Identifier | KEGG Reaction |
| Spermine | 3-aminopropanal | Spermidine | EC 1.5.3.16, EC 1.5.3.17 | R09076 |
| Spermidine | 3-aminopropanal | Putrescine | EC 1.5.3.17 | R09077 |
| 4-aminobutanal | 1,3-diaminopropane | EC 1.5.3.14 | R01914 | |
| N1-acetylspermine | N-(3-acetamidopropyl)-4-aminobutanal | 1,3-diaminopropane | EC 1.5.3.15 | NA |
| 3-acetamidopropanal | Spermidine | EC 1.5.3.13, EC 1.5.3.17 | R03899 | |
| N1,N12-diacetylspermine | 3-acetamidopropanal | N1-acetylspermidine | EC 1.5.3.13 | NA |
| N1-acetylspermidine | 3-acetamidopropanal | Putrescine | EC 1.5.3.13, EC 1.5.3.17 | R09074 |
| N8-acetylspermidine | 4-acetamidobutanal | 1,3-diaminopropane | EC 1.5.3.15 | R09075 |
Structure and Mechanism
Structures of PAOs from corn, brewer's yeast, and mice contain a substrate-binding domain and an FAD-binding domain that secures the FAD cofactor non-covalently.[2][3][4] The active site is located at the interface of these domains.
Active sites in PAOs vary, but some features are essential. The most strictly-conserved active site amino acid codons in PAO genes are a K residue at position 300 and an aromatic residue (F, Y, or H) at position 403 (numbers refer to homologous positions in the sequence of ZmPAO1, a PAO found in corn).[1] K300 hydrogen-bonds to a water molecule, which also hydrogen-bonds to the catalytic N5 nitrogen atom of FAD. In corn, this complex modulates redox potential and reoxidation rate of the cofactor and may be involved in stabilizing the reduced cofactor or imine hydrolysis.[6] The electron-dense aromatic ring at position 403 may interact with amino groups in the substrate.[7] Weak interactions with other more variable active site residues are involved in positioning the substrates.[8]
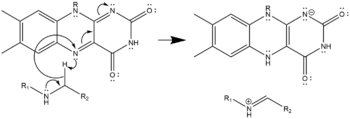
In the active site, FAD oxidizes the secondary amine to an imine, which water subsequently hydrolyzes, yielding an amino-aldehyde and a primary amine. Molecular oxygen reoxidizes the FAD, generating hydrogen peroxide.
- Three possible mechanisms of polyamine oxidation by FAD based on the structure of mouse N1-Acetylspermine Oxidase;[9] the secondary amine oxidized must be in the charge-neutral, deprotonated state. This oxidation may proceed by hydride transfer (left), radical abstraction (center), or nucleophilic attack with a covalently-bonded cofactor-substrate intermediate (right). All three are equally plausible given the structure and none has been confirmed as the true mechanism.

Metabolism
PAOs are central to polyamine catabolism. Different organic reaction products result in different metabolites with different fates. Because all PAO reactions release hydrogen peroxide, PAOs are tied to the metabolism of reactive oxygen species, which overlaps with pathways of programmed cell death.[11]
Polyamine catabolism is often upregulated in mammalian tumor cells. Molecules downstream of polyamine oxidation play roles in cell proliferation. Spermidine is a precursor to hypusine, which activates eukaryotic initiation factor 5A isoform 1 (eIF5A) that enables translation of mRNA. Putrescine has effects on mTOR complex 1 and eukaryotic translation initiation factor 4E (eIF4FE). Because of this, multiple anticancer drugs in various stages of clinical trial target PAOs.[12]
In plants, polyamine catabolism is tied closely to stress responses and fruit ripening.[13]
References
- ↑ 1.0 1.1 Salvi, Daniele; Tavladoraki, Paraskevi (2020-10-20). "The tree of life of polyamine oxidases" (in en). Scientific Reports 10 (1): 17858. doi:10.1038/s41598-020-74708-3. ISSN 2045-2322. PMID 33082384. Bibcode: 2020NatSR..1017858S.
- ↑ Binda, Claudia; Coda, Alessandro; Angelini, Riccardo; Federico, Rodolfo; Ascenzi, Paolo; Mattevi, Andrea (1999-03-15). "A 30 Å long U-shaped catalytic tunnel in the crystal structure of polyamine oxidase" (in en). Structure 7 (3): 265–276. doi:10.1016/S0969-2126(99)80037-9. ISSN 0969-2126. PMID 10368296.
- ↑ Huang, Qingqiu; Liu, Qun; Hao, Quan (2005-05-13). "Crystal structures of Fms1 and its complex with spermine reveal substrate specificity". Journal of Molecular Biology 348 (4): 951–959. doi:10.1016/j.jmb.2005.03.008. ISSN 0022-2836. PMID 15843025. https://pubmed.ncbi.nlm.nih.gov/15843025/.
- ↑ Sjögren, Tove; Wassvik, Carola M.; Snijder, Arjan; Aagaard, Anna; Kumanomidou, Taichi; Barlind, Louise; Kaminski, Tim P.; Kashima, Akiko et al. (2017-01-24). "The Structure of Murine N1-Acetylspermine Oxidase Reveals Molecular Details of Vertebrate Polyamine Catabolism". Biochemistry 56 (3): 458–467. doi:10.1021/acs.biochem.6b01140. ISSN 0006-2960. PMID 28029774. https://doi.org/10.1021/acs.biochem.6b01140.
- ↑ Bank, RCSB Protein Data (in en). 3D View: 1H84. https://www.rcsb.org/3d-view/ngl/1h84.
- ↑ Fiorillo, Annarita; Federico, Rodolfo; Polticelli, Fabio; Boffi, Alberto; Mazzei, Franco; Fusco, Massimo Di; Ilari, Andrea; Tavladoraki, Paraskevi (2011). "The structure of maize polyamine oxidase K300M mutant in complex with the natural substrates provides a snapshot of the catalytic mechanism of polyamine oxidation" (in en). The FEBS Journal 278 (5): 809–821. doi:10.1111/j.1742-4658.2010.08000.x. ISSN 1742-4658. PMID 21205212. https://onlinelibrary.wiley.com/doi/abs/10.1111/j.1742-4658.2010.08000.x.
- ↑ Binda, C.; Mattevi, A.; Edmondson, D. E. (2002-07-05). "Structure-Function Relationships in Flavoenzyme-dependent Amine Oxidations: A COMPARISON OF POLYAMINE OXIDASE AND MONOAMINE OXIDASE" (in en). Journal of Biological Chemistry 277 (27): 23973–23976. doi:10.1074/jbc.R200005200. ISSN 0021-9258. PMID 12015330.
- ↑ Binda, Claudia; Angelini, Riccardo; Federico, Rodolfo; Ascenzi, Paolo; Mattevi, Andrea (2001-03-01). "Structural Bases for Inhibitor Binding and Catalysis in Polyamine Oxidase". Biochemistry 40 (9): 2766–2776. doi:10.1021/bi002751j. ISSN 0006-2960. PMID 11258887. https://doi.org/10.1021/bi002751j.
- ↑ Sjögren, Tove; Wassvik, Carola M.; Snijder, Arjan; Aagaard, Anna; Kumanomidou, Taichi; Barlind, Louise; Kaminski, Tim P.; Kashima, Akiko et al. (2017-01-24). "The Structure of Murine N1-Acetylspermine Oxidase Reveals Molecular Details of Vertebrate Polyamine Catabolism". Biochemistry 56 (3): 458–467. doi:10.1021/acs.biochem.6b01140. ISSN 0006-2960. PMID 28029774. https://doi.org/10.1021/acs.biochem.6b01140.
- ↑ Romero, Elvira; Gómez Castellanos, J. Rubén; Gadda, Giovanni; Fraaije, Marco W.; Mattevi, Andrea (2018-02-28). "Same Substrate, Many Reactions: Oxygen Activation in Flavoenzymes" (in en). Chemical Reviews 118 (4): 1742–1769. doi:10.1021/acs.chemrev.7b00650. ISSN 0009-2665. PMID 29323892.
- ↑ Moschou, Panagiotis N.; Roubelakis-Angelakis, Kalliopi A. (2014-03-01). "Polyamines and programmed cell death". Journal of Experimental Botany 65 (5): 1285–1296. doi:10.1093/jxb/ert373. ISSN 0022-0957. PMID 24218329.
- ↑ Casero, Robert A.; Murray Stewart, Tracy; Pegg, Anthony E. (November 2018). "Polyamine metabolism and cancer: treatments, challenges and opportunities". Nature Reviews. Cancer 18 (11): 681–695. doi:10.1038/s41568-018-0050-3. ISSN 1474-1768. PMID 30181570.
- ↑ Wang, Wei; Paschalidis, Konstantinos; Feng, Jian-Can; Song, Jie; Liu, Ji-Hong (2019). "Polyamine Catabolism in Plants: A Universal Process With Diverse Functions". Frontiers in Plant Science 10: 561. doi:10.3389/fpls.2019.00561. ISSN 1664-462X. PMID 31134113.
 |