Biology:Polyclonal B cell response

Polyclonal B cell response is a natural mode of immune response exhibited by the adaptive immune system of mammals. It ensures that a single antigen is recognized and attacked through its overlapping parts, called epitopes, by multiple clones of B cell.[1][2]
In the course of normal immune response, parts of pathogens (e.g. bacteria) are recognized by the immune system as foreign (non-self), and eliminated or effectively neutralized to reduce their potential damage. Such a recognizable substance is called an antigen. The immune system may respond in multiple ways to an antigen; a key feature of this response is the production of antibodies by B cells (or B lymphocytes) involving an arm of the immune system known as humoral immunity. The antibodies are soluble and do not require direct cell-to-cell contact between the pathogen and the B-cell to function.
Antigens can be large and complex substances, and any single antibody can only bind to a small, specific area on the antigen. Consequently, an effective immune response often involves the production of many different antibodies by many different B cells against the same antigen. Hence the term "polyclonal", which derives from the words poly, meaning many, and clones from Greek klōn, meaning sprout or twig;[3][4][5] a clone is a group of cells arising from a common "mother" cell. The antibodies thus produced in a polyclonal response are known as polyclonal antibodies. The heterogeneous polyclonal antibodies are distinct from monoclonal antibody molecules, which are identical and react against a single epitope only, i.e., are more specific.
Although the polyclonal response confers advantages on the immune system, in particular, greater probability of reacting against pathogens, it also increases chances of developing certain autoimmune diseases resulting from the reaction of the immune system against native molecules produced within the host.
Humoral response to infection
Diseases which can be transmitted from one organism to another are known as infectious diseases, and the causative biological agent involved is known as a pathogen. The process by which the pathogen is introduced into the body is known as inoculation,[note 1][6] and the organism it affects is known as a biological host. When the pathogen establishes itself in a step known as colonization,[7] it can result in an infection,[7] consequently harming the host directly or through the harmful substances called toxins it can produce.[7] This results in the various symptoms and signs characteristic of an infectious disease like pneumonia or diphtheria.
Countering the various infectious diseases is very important for the survival of the susceptible organism, in particular, and the species, in general. This is achieved by the host by eliminating the pathogen and its toxins or rendering them nonfunctional. The collection of various cells, tissues and organs that specializes in protecting the body against infections is known as the immune system. The immune system accomplishes this through direct contact of certain white blood cells with the invading pathogen involving an arm of the immune system known as the cell-mediated immunity, or by producing substances that move to sites distant from where they are produced, "seek" the disease-causing cells and toxins by specifically[note 2] binding with them, and neutralize them in the process–known as the humoral arm of the immune system. Such substances are known as soluble antibodies and perform important functions in countering infections.[note 3][8]
- Types of White blood cells (WBCs)
B cell response

Antibodies serve various functions in protecting the host against the pathogen. Their soluble forms which carry out these functions are produced by plasma B cells, a type of white blood cell. This production is tightly regulated and requires the activation of B cells by activated T cells (another type of white blood cell), which is a sequential procedure. The major steps involved are:[9]
- Specific or nonspecific recognition of the pathogen (because of its antigens) with its subsequent engulfing by B cells or macrophages. This activates the B cell only partially.
- Antigen processing.
- Antigen presentation.
- Activation of the T helper cells by antigen-presenting cells.
- Co-stimulation of the B cell by activated T cell resulting in its complete activation.
- Proliferation[note 4] of B cells with resultant production of soluble antibodies.

Recognition of pathogens
Pathogens synthesize proteins that can serve as "recognizable" antigens; they may express the molecules on their surface or release them into the surroundings (body fluids). What makes these substances recognizable is that they bind very specifically and somewhat strongly to certain host proteins called antibodies. The same antibodies can be anchored to the surface of cells of the immune system, in which case they serve as receptors, or they can be secreted in the blood, known as soluble antibodies. On a molecular scale, the proteins are relatively large, so they cannot be recognized as a whole; instead, their segments, called epitopes, can be recognized.[1] An epitope comes in contact with a very small region (of 15–22 amino acids) of the antibody molecule; this region is known as the paratope.[1] In the immune system, membrane-bound antibodies are the B-cell receptor (BCR). Also, while the T-cell receptor is not biochemically classified as an antibody, it serves a similar function in that it specifically binds to epitopes complexed with major histocompatibility complex (MHC) molecules.[note 5][10] The binding between a paratope and its corresponding antigen is very specific, owing to its structure, and is guided by various noncovalent bonds, not unlike the pairing of other types of ligands (any atom, ion or molecule that binds with any receptor with at least some degree of specificity and strength). The specificity of binding does not arise out of a rigid lock and key type of interaction, but rather requires both the paratope and the epitope to undergo slight conformational changes in each other's presence.[11]
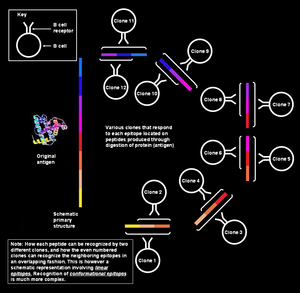
Specific recognition of epitope by B cells

In figure at left, the various segments that form the epitope have been shown to be continuously collinear, meaning that they have been shown as sequential; however, for the situation being discussed here (i.e., the antigen recognition by the B cell), this explanation is too simplistic. Such epitopes are known as sequential or linear epitopes, as all the amino acids on them are in the same sequence (line). This mode of recognition is possible only when the peptide is small (about six to eight amino acids long),[1] and is employed by the T cells (T lymphocytes).
However, the B memory/naive cells recognize intact proteins present on the pathogen surface.[note 6] In this situation, the protein in its tertiary structure is so greatly folded that some loops of amino acids come to lie in the interior of the protein, and the segments that flank them may lie on the surface. The paratope on the B cell receptor comes in contact only with those amino acids that lie on the surface of the protein. The surface amino acids may actually be discontinuous in the protein's primary structure, but get juxtaposed owing to the complex protein folding patterns (as in the adjoining figure). Such epitopes are known as conformational epitopes and tend to be longer (15–22 amino acid residues) than the linear epitopes.[1] Likewise, the antibodies produced by the plasma cells belonging to the same clone would bind to the same conformational epitopes on the pathogen proteins.[12][13][14][15]
The binding of a specific antigen with corresponding BCR molecules results in increased production of the MHC-II molecules. This assumes significance as the same does not happen when the same antigen would be internalized by a relatively nonspecific process called pinocytosis, in which the antigen with the surrounding fluid is "drunk" as a small vesicle by the B cell.[16] Hence, such an antigen is known as a nonspecific antigen and does not lead to activation of the B cell, or subsequent production of antibodies against it.
Nonspecific recognition by macrophages
Macrophages and related cells employ a different mechanism to recognize the pathogen. Their receptors recognize certain motifs present on the invading pathogen that are very unlikely to be present on a host cell. Such repeating motifs are recognized by pattern recognition receptors (PRRs) like the toll-like receptors (TLRs) expressed by the macrophages.[1][17] Since the same receptor could bind to a given motif present on surfaces of widely disparate microorganisms, this mode of recognition is relatively nonspecific, and constitutes an innate immune response.
Antigen processing
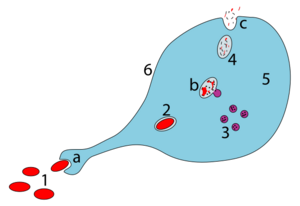
After recognizing an antigen, an antigen-presenting cell such as the macrophage or B lymphocyte engulfs it completely by a process called phagocytosis. The engulfed particle, along with some material surrounding it, forms the endocytic vesicle (the phagosome), which fuses with lysosomes. Within the lysosome, the antigen is broken down into smaller pieces called peptides by proteases (enzymes that degrade larger proteins). The individual peptides are then complexed with major histocompatibility complex class II (MHC class II) molecules located in the lysosome – this method of "handling" the antigen is known as the exogenous or endocytic pathway of antigen processing in contrast to the endogenous or cytosolic pathway,[17][18][19] which complexes the abnormal proteins produced within the cell (e.g. under the influence of a viral infection or in a tumor cell) with MHC class I molecules.
An alternate pathway of endocytic processing had also been demonstrated wherein certain proteins like fibrinogen and myoglobin can bind as a whole to MHC-II molecules after they are denatured and their disulfide bonds are reduced (breaking the bond by adding hydrogen atoms across it). The proteases then degrade the exposed regions of the protein-MHC II-complex.[19]
Antigen presentation
After the processed antigen (peptide) is complexed to the MHC molecule, they both migrate together to the cell membrane, where they are exhibited (elaborated) as a complex that can be recognized by the CD 4+ (T helper cell) – a type of white blood cell.[note 7][20] This is known as antigen presentation. However, the epitopes (conformational epitopes) that are recognized by the B cell prior to their digestion may not be the same as that presented to the T helper cell. Additionally, a B cell may present different peptides complexed to different MHC-II molecules.[16]
T helper cell stimulation
The CD 4+ cells through their T cell receptor-CD3 complex recognize the epitope-bound MHC II molecules on the surface of the antigen presenting cells, and get 'activated'. Upon this activation, these T cells proliferate and differentiate into Th1 or Th2 cells.[16][21] This makes them produce soluble chemical signals that promote their own survival. However, another important function that they carry out is the stimulation of B cell by establishing direct physical contact with them.[10]
Co-stimulation of B cell by activated T helper cell
Complete stimulation of T helper cells requires the B7 molecule present on the antigen presenting cell to bind with CD28 molecule present on the T cell surface (in close proximity with the T cell receptor).[10] Likewise, a second interaction between the CD40 ligand or CD154 (CD40L) present on T cell surface and CD40 present on B cell surface, is also necessary.[21] The same interactions that stimulate the T helper cell also stimulate the B cell, hence the term costimulation. The entire mechanism ensures that an activated T cell only stimulates a B cell that recognizes the antigen containing the same epitope as recognized by the T cell receptor of the "costimulating" T helper cell. The B cell gets stimulated, apart from the direct costimulation, by certain growth factors, viz., interleukins 2, 4, 5, and 6 in a paracrine fashion. These factors are usually produced by the newly activated T helper cell.Cite error: Closing </ref> missing for <ref> tag the rest survive as memory B cells. So, when the naive cells belonging to a particular clone encounter their specific antigen to give rise to the plasma cells, and also leave a few memory cells, this is known as the primary immune response. In the course of proliferation of this clone, the B cell receptor genes can undergo frequent (one in every two cell divisions)[8] mutations in the genes coding for paratopes of antibodies. These frequent mutations are termed somatic hypermutation. Each such mutation alters the epitope-binding ability of the paratope slightly, creating new clones of B cells in the process. Some of the newly created paratopes bind more strongly to the same epitope (leading to the selection of the clones possessing them), which is known as affinity maturation.[note 8][8][21] Other paratopes bind better to epitopes that are slightly different from the original epitope that had stimulated proliferation. Variations in the epitope structure are also usually produced by mutations in the genes of pathogen coding for their antigen. Somatic hypermutation, thus, makes the B cell receptors and the soluble antibodies in subsequent encounters with antigens, more inclusive in their antigen recognition potential of altered epitopes, apart from bestowing greater specificity for the antigen that induced proliferation in the first place. When the memory cells get stimulated by the antigen to produce plasma cells (just like in the clone's primary response), and leave even more memory cells in the process, this is known as a secondary immune response,[21] which translates into greater numbers of plasma cells and faster rate of antibody production lasting for longer periods. The memory B cells produced as a part of secondary response recognize the corresponding antigen faster and bind more strongly with it (i.e., greater affinity of binding) owing to affinity maturation. The soluble antibodies produced by the clone show a similar enhancement in antigen binding.[21]
Basis of polyclonality
Responses are polyclonal in nature as each clone somewhat specializes in producing antibodies against a given epitope, and because, each antigen contains multiple epitopes, each of which in turn can be recognized by more than one clone of B cells. To be able to react to innumerable antigens, as well as multiple constituent epitopes, the immune system requires the ability to recognize a very great number of epitopes in all, i.e., there should be a great diversity of B cell clones.
Clonality of B cells
Memory and naïve B cells normally exist in relatively small numbers. As the body needs to be able to respond to a large number of potential pathogens, it maintains a pool of B cells with a wide range of specificities.[17] Consequently, while there is almost always at least one B (naive or memory) cell capable of responding to any given epitope (of all that the immune system can react against), there are very few exact duplicates. However, when a single B cell encounters an antigen to which it can bind, it can proliferate very rapidly.[21] Such a group of cells with identical specificity towards the epitope is known as a clone, and is derived from a common "mother" cell. All the "daughter" B cells match the original "mother" cell in their epitope specificity, and they secrete antibodies with identical paratopes. These antibodies are monoclonal antibodies, since they derive from clones of the same parent cell. A polyclonal response is one in which clones of multiple B cells react to the same antigen.
Single antigen contains multiple overlapping epitopes

A single antigen can be thought of as a sequence of multiple overlapping epitopes. Many unique B cell clones may be able to bind to the individual epitopes. This imparts even greater multiplicity to the overall response.[3] All of these B cells can become activated and produce large colonies of plasma cell clones, each of which can secrete up to 1000 antibody molecules against each epitope per second.[21]
Multiple clones recognize single epitope
In addition to different B cells reacting to different epitopes on the same antigen, B cells belonging to different clones may also be able to react to the same epitope. An epitope that can be attacked by many different B cells is said to be highly immunogenic. In these cases, the binding affinities for respective epitope-paratope pairs vary, with some B cell clones producing antibodies that bind strongly to the epitope, and others producing antibodies that bind weakly.[1]
Clonal selection
The clones that bind to a particular epitope with greater strength are more likely to be selected for further proliferation in the germinal centers of the follicles in various lymphoid tissues like the lymph nodes. This is not unlike natural selection: clones are selected for their fitness to attack the epitopes (strength of binding) on the encountered pathogen.[22] What makes the analogy even stronger is that the B lymphocytes have to compete with each other for signals that promote their survival in the germinal centers.
Diversity of B cell clones
Although there are many diverse pathogens, many of which are constantly mutating, it is a surprise that a majority of individuals remain free of infections. Thus, maintenance of health requires the body to recognize all pathogens (antigens they present or produce) likely to exist. This is achieved by maintaining a pool of immensely large (about 109) clones of B cells, each of which reacts against a specific epitope by recognizing and producing antibodies against it. However, at any given time very few clones actually remain receptive to their specific epitope. Thus, approximately 107 different epitopes can be recognized by all the B cell clones combined.[21] Moreover, in a lifetime, an individual usually requires the generation of antibodies against very few antigens in comparison with the number that the body can recognize and respond against.[21]
Significance of the phenomenon
Increased probability of recognizing any antigen
If an antigen can be recognized by more than one component of its structure, it is less likely to be "missed" by the immune system.[note 9] Mutation of pathogenic organisms can result in modification of antigen—and, hence, epitope—structure. If the immune system "remembers" what the other epitopes look like, the antigen, and the organism, will still be recognized and subjected to the body's immune response. Thus, the polyclonal response widens the range of pathogens that can be recognized.[23]
Limitation of immune system against rapidly mutating viruses

Many viruses undergo frequent mutations that result in changes in amino acid composition of their important proteins. Epitopes located on the protein may also undergo alterations in the process. Such an altered epitope binds less strongly with the antibodies specific to the unaltered epitope that would have stimulated the immune system. This is unfortunate because somatic hypermutation does give rise to clones capable of producing soluble antibodies that would have bound the altered epitope avidly enough to neutralize it. But these clones would consist of naive cells which are not allowed to proliferate by the weakly binding antibodies produced by the priorly stimulated clone. This doctrine is known as the original antigenic sin.[21] This phenomenon comes into play particularly in immune responses against influenza, dengue and HIV viruses.[24] This limitation, however, is not imposed by the phenomenon of polyclonal response, but rather, against it by an immune response that is biased in favor of experienced memory cells against the "novice" naive cells.
Increased chances of autoimmune reactions
In autoimmunity the immune system wrongly recognizes certain native molecules in the body as foreign (self-antigen), and mounts an immune response against them. Since these native molecules, as normal parts of the body, will naturally always exist in the body, the attacks against them can get stronger over time (akin to secondary immune response). Moreover, many organisms exhibit molecular mimicry, which involves showing those antigens on their surface that are antigenically similar to the host proteins. This has two possible consequences: first, either the organism will be spared as a self antigen; or secondly, that the antibodies produced against it will also bind to the mimicked native proteins. The antibodies will attack the self-antigens and the tissues harboring them by activating various mechanisms like the complement activation and antibody-dependent cell-mediated cytotoxicity. Hence, wider the range of antibody-specificities, greater the chance that one or the other will react against self-antigens (native molecules of the body).[25][26]
Difficulty in producing monoclonal antibodies
Monoclonal antibodies are structurally identical immunoglobulin molecules with identical epitope-specificity (all of them bind with the same epitope with same affinity) as against their polyclonal counterparts which have varying affinities for the same epitope. They are usually not produced in a natural immune response, but only in diseased states like multiple myeloma, or through specialized laboratory techniques. Because of their specificity, monoclonal antibodies are used in certain applications to quantify or detect the presence of substances (which act as antigen for the monoclonal antibodies), and for targeting individual cells (e.g. cancer cells). Monoclonal antibodies find use in various diagnostic modalities (see: western blot and immunofluorescence) and therapies—particularly of cancer and diseases with autoimmune component. But, since virtually all responses in nature are polyclonal, it makes production of immensely useful monoclonal antibodies less straightforward.[8]
History
The first evidence of presence of a neutralizing substance in the blood that could counter infections came when Emil von Behring along with Kitasato Shibasaburō in 1890 developed effective serum against diphtheria. This they did by transferring serum produced from animals immunized against diphtheria to animals suffering from it. Transferring the serum thus could cure the infected animals. Behring was awarded the Nobel Prize for this work in 1901.[27]
At this time though the chemical nature of what exactly in the blood conferred this protection was not known. In a few decades to follow, it was shown that the protective serum could neutralize and precipitate toxins, and clump bacteria. All these functions were attributed to different substances in the serum, and named accordingly as antitoxin, precipitin and agglutinin.[17] That all the three substances were one entity (gamma globulins) was demonstrated by Elvin A. Kabat in 1939. In the preceding year Kabat had demonstrated the heterogeneity of antibodies through ultracentrifugation studies of horses' sera.[28]
Until this time, cell-mediated immunity and humoral immunity were considered to be contending theories to explain effective immune response, but the former lagged behind owing to lack of advanced techniques.[17] Cell-mediated immunity got an impetus in its recognition and study when in 1942, Merrill Chase successfully transferred immunity against tuberculosis between pigs by transferring white blood cells.[17][29]

It was later shown in 1948 by Astrid Fagraeus in her doctoral thesis that the plasma B cells are specifically involved in antibody production.[30] The role of lymphocytes in mediating both cell-mediated and humoral responses was demonstrated by James Gowans in 1959.[29]
In order to account for the wide range of antigens the immune system can recognize, Paul Ehrlich in 1900 had hypothesized that preexisting "side chain receptors" bind a given pathogen, and that this interaction induces the cell exhibiting the receptor to multiply and produce more copies of the same receptor. This theory, called the selective theory was not proven for next five decades, and had been challenged by several instructional theories which were based on the notion that an antibody would assume its effective structure by folding around the antigen.[17] In the late 1950s however, the works of three scientists—Jerne, Talmage and Burnet (who largely modified the theory)—gave rise to the clonal selection theory, which proved all the elements of Ehrlich's hypothesis except that the specific receptors that could neutralize the agent were soluble and not membrane-bound.[17][29]
The clonal selection theory was proved correct when Sir Gustav Nossal showed that each B cell always produces only one antibody.[31]
In 1974, the role of MHC in antigen presentation was demonstrated by Rolf Zinkernagel and Peter C. Doherty.[29]
See also
- Polyclonal antibodies
- Antigen processing
- Antiserum, a polyclonal antibody preparation used to treat envenomation
Notes
- ↑ The term "inoculation" is usually used in context of active immunization, i.e., deliberately introducing the antigenic substance into the host's body. But in many discussions of infectious diseases, it is not uncommon to use the term to imply a spontaneous (that is, without human intervention) event resulting in introduction of the causative organism into the body, say ingesting water contaminated with Salmonella typhi—the causative organism for typhoid fever. In such cases the causative organism itself is known as the inoculum, and the number of organisms introduced as the "dose of inoculum".
- ↑ Specificity implies that two different pathogens will be actually viewed as two distinct entities, and countered by different antibody molecules.
- ↑ Actions of antibodies:
- Coating the pathogen, preventing it from adhering to the host cell, and thus preventing colonization
- Precipitating (making the particles "sink" by attaching to them) the soluble antigens and promoting their clearance by other cells of immune system from the various tissues and blood
- Coating the microorganisms to attract cells that can engulf the pathogen. This is known as opsonization. Thus the antibody acts as an opsonin. The process of engulfing is known as phagocytosis (literally, cell eating)
- Activating the complement system, which most importantly pokes holes into the pathogen's outer covering (its cell membrane), killing it in the process
- Marking up host cells infected by viruses for destruction in a process known as Antibody-dependent cell-mediated cytotoxicity (ADCC)
- ↑ Proliferation in this context means multiplication by cell division and differentiation
- ↑ The major histocompatibility complex is a gene region on the DNA that codes for the synthesis of Major histocompatibility class I molecule, Major histocompatibility class II molecule and other proteins involved in the function of complement system (MHC class III). The first two products are important in antigen presentation. MHC-compatibility is a major consideration in organ transplantation, and in humans is also known as the human leukocyte antigen (HLA).
- ↑ Here, intact implies that the undigested protein is recognized, and not that the paratope on B cell receptor comes in contact with the whole protein structure at the same time; the paratope will still contact only a restricted portion of the antigen exposed on its surface.
- ↑ There are many types of white blood cells. The common way of classifying them is according to their appearance under the light microscope after they are stained by chemical dyes. But with advancing technology newer methods of classification has emerged. One of the methods employs the use of monoclonal antibodies, which can bind specifically to each type of cell. Moreover, the same type of white blood cell would express molecules typical to it on its cell membrane at various stages of development. The monoclonal antibodies that can specifically bind with a particular surface molecule would be regarded as one cluster of differentiation (CD). Any monoclonal antibody or group of monoclonal antibodies that does not react with known surface molecules of lymphocytes, but rather to a yet-unrecognized surface molecule would be clubbed as a new cluster of differentiation and numbered accordingly. Each cluster of differentiation is abbreviated as "CD", and followed by a number (usually indicating the order of discovery). So, a cell possessing a surface molecule (called ligand) that binds specifically to cluster of differentiation 4 would be known as CD4+ cell. Likewise, a CD8+ cell is one that would possess the CD8 ligand and bind to CD8 monoclonal antibodies.
- ↑ Affinity roughly translates as attraction from Latin. See also: Definition of Affinity from Online Etymology Dictionary and Definition of Affinity from TheFreeDictionary by Farlex
- ↑ Analogically, if in a crowded place, one is supposed to recognize a person, it is better to know as many physical features as possible. If you know the person only by the hairstyle, there is a chance of overlooking the person if that changes. Whereas, if apart from the hairstyle, if you also happen to know the facial features and what the person will wear on a particular day, it becomes much more unlikely that you will miss that person.
References
- ↑ 1.0 1.1 1.2 1.3 1.4 1.5 1.6 1.7 1.8 Goldsby, Richard; Kindt, TJ; Osborne, BA; Janis Kuby (2003). "Antigens (Chapter 3)". Immunology (Fifth ed.). New York: W. H. Freeman and Company. pp. 57–75. ISBN 978-0716749479. https://archive.org/details/immunology00gold_0/page/57.
- ↑ "Definition of Polyclonal from MedicineNet.com". Webster's New World Medical Dictionary. http://www.medterms.com/script/main/art.asp?articlekey=20127.
- ↑ 3.0 3.1 Frank, Steven A. (2002). "Specificity and Cross-Reactivity (Chapter 4)". Immunology and Evolution of Infectious Disease. Princeton University. pp. 33–56. ISBN 978-0691095950. https://www.ncbi.nlm.nih.gov/bookshelf/br.fcgi?book=infdis&part=A22#A23. Retrieved 2008-06-23.
- ↑ "Etymology of "clone"". Online etymology dictionary. http://www.etymonline.com/index.php?search=clone&searchmode=none.
- ↑ Bansal, R.K. (2005). "Reproductive Cloning-An Act Of Human Rights Violation". Journal of Indian Association of Forensic Medicine 27 (3): 971–973. http://medind.nic.in/jal/t05/i3/jalt05i3p180.pdf. Retrieved 2008-06-23.
- ↑ "Definition of inoculation". TheFreeDictionary.com (citing Dorland's Medical Dictionary for Health Consumers. © 2007 by Saunders, an imprint of Elsevier, Inc.). http://medical-dictionary.thefreedictionary.com/inoculation.
- ↑ 7.0 7.1 7.2 Pier, Gerald B. (2005). "Molecular mechanisms of microbial pathogenesis (Chapter 105)". Harrison's Principles of Internal Medicine. 1 (Sixteenth ed.). McGraw-Hill. pp. 700. ISBN 978-0-07-123983-7.
- ↑ 8.0 8.1 8.2 8.3 8.4 Goldsby (2007). "Organization and Expression of Immunoglobulin Genes (Chapter 5)". Immunology (Fifth ed.). New York. pp. 105–136. ISBN 978-0-7167-6764-0.
- ↑ Nairn, Roderick (2004). "Immunology (Chapter 8)". in Geo F. Brooks. Jawetz, Melnick, & Adelberg's Medical Microbiology (Twenty-Third Edition International ed.). Lange publications/McGraw-Hill. pp. 133–135, 138–139. ISBN 978-0-07-123983-7.
- ↑ 10.0 10.1 10.2 Goldsby (2007). "T-Cell Maturation, Activation and Differentiation (Chapter 10)". Immunology (Fifth ed.). W.H. Freeman. pp. 221–246. ISBN 978-0-7167-6764-0.
- ↑ Nair, Deepak; Singh Kavita; Siddiqui Zaved; Nayak Bishnu; Rao Kanury; Salunke Dinakar (2002-01-09). "Epitope Recognition by Diverse Antibodies Suggests Conformational Convergence in an Antibody Response". The Journal of Immunology 168 (5): 2371–2382. doi:10.4049/jimmunol.168.5.2371. PMID 11859128. http://www.jimmunol.org/cgi/reprint/168/5/2371.pdf. Retrieved 2008-05-03.
- ↑ "Immunochemical Applications". Technical Tips. EMD biosciences. http://www.emdbiosciences.com/html/CBC/technical-tips-immunochemical-applications.html#TechTip8.
- ↑ Davis, Cheryl. "Antigens". Biology course. Western Kentucky University. http://bioweb.wku.edu/courses/biol328/antigens.html.
- ↑ Ceri, Howard. "Antigens". Immunology course. University of Calgary. https://www.ucalgary.ca/~ceri/cmmb427prot/ANTIGEN.html.
- ↑ Khudyakov, Yury; Howard A. Fields (2002). Artificial DNA: Methods and Applications. Florida: CRC Press. pp. 227. ISBN 978-0-8493-1426-1. https://books.google.com/books?id=dexRnDtLlWUC&q=nonsequential+epitopes+amino+acid&pg=PA227.
- ↑ 16.0 16.1 16.2 Myers, CD (1991). "Role of B cell antigen processing and presentation in the humoral immune response". The FASEB Journal 5 (11): 2547–2553. doi:10.1096/fasebj.5.11.1907935. PMID 1907935. http://www.fasebj.org/cgi/reprint/5/11/2547.pdf. Retrieved 2008-06-20.
- ↑ 17.0 17.1 17.2 17.3 17.4 17.5 17.6 17.7 Goldsby (2007). "Overview of the Immune System (Chapter 1)". Immunology (Fifth ed.). W.H. Freeman. pp. 1–23. ISBN 978-0-7167-6764-0.
- ↑ Goldsby (2007). "Antigen Processing and Presentation (Chapter 8)". Immunology (Fifth ed.). W.H. Freeman. pp. 188–194. ISBN 978-0-7167-6764-0.
- ↑ 19.0 19.1 Ojcius, DM; L Gapin; JM Kanellopoulos; P Kourilsky (September 1994). "Is antigen processing guided by major histocompatibility complex molecules?". The FASEB Journal 8 (5): 974–978. doi:10.1096/fasebj.8.12.8088463. PMID 8088463. http://www.fasebj.org/cgi/reprint/8/12/974.pdf. Retrieved 2008-06-20.
- ↑ Goldsby (2007). "Cells and Organs of the Immune System (Chapter 2)". Immunology (Fifth ed.). W.H. Freeman. pp. 24–56. ISBN 978-0-7167-6764-0.
- ↑ 21.00 21.01 21.02 21.03 21.04 21.05 21.06 21.07 21.08 21.09 21.10 Goldsby (2007). "B-Cell Generation, Activation and Differentiation (Chapter 11)". Immunology (Fifth ed.). New York. pp. 247–275. ISBN 978-0-7167-6764-0.
- ↑ Cziko, Gary (1995). "The Immune System: Selection by the Enemy". Without Miracles: Universal Selection Theory and the Second Darwinian Revolution (Fifth ed.). Massachusetts: MIT Press. pp. 39–48. ISBN 978-0-262-03232-2. http://faculty.ed.uiuc.edu/g-cziko/wm/04.html#Heading5. Retrieved 2008-05-12.
- ↑ Greener, Mark (2005-02-14). "Monoclonal antibodies (MAbs) turn 30". The Scientist 19 (3): 14. http://www.vetscite.org/publish/items/002093/index.html#. Retrieved 2008-06-06.
- ↑ Deem, Michael. "Michael W. Deem". Official Web Page. Rice University. http://www.mwdeem.rice.edu/mwdeem/.
- ↑ Granholm, Norman; Tito Cavallo (1992). "Autoimmunity, Polyclonal B-Cell Activation and Infection (abstract)". Lupus 1 (2): 63–74. doi:10.1177/096120339200100203. PMID 1301966.
- ↑ "Polyclonal B cell activation in infections: infectious agents' devilry or defense mechanism of the host? (abstract)". Journal of Leukocyte Biology 82 (5): 1027–1032. 2007. doi:10.1189/jlb.0407214. PMID 17615380.
- ↑ "Emil von Behring: The Founder of Serum Therapy". Nobel Prize in Medicine. http://nobelprize.org/nobel_prizes/medicine/articles/behring/index.html.
- ↑ Mage, Rose G.; Ten Feizi. "Elvin A. Kabat". Biographical memoirs. http://www.nap.edu/readingroom/books/biomems/ekabat.html.
- ↑ 29.0 29.1 29.2 29.3 Greenberg, Steven. "A Concise History of Immunology". http://www.columbia.edu/itc/hs/medical/pathophys/immunology/readings/ConciseHistoryImmunology.pdf.
- ↑ "MTC News". Karolinska Institutet. http://ki.se/content/1/c6/02/28/99/get_pdf-17.php.pdf.
- ↑ Turner, Stephen (October 2007). "One POWERFUL Idea". Australasian Science. http://www.control.com.au/bi2007/289Burnet.pdf.
Further reading
- Goldsby, Richard; Kindt, TJ; Osborne, BA; Janis Kuby (2003). Immunology (Fifth ed.). New York: W. H. Freeman and Company. ISBN 978-0-7167-4947-9. https://archive.org/details/immunology00gold_0.
- Kishiyama, Jeffery L. (2006). "Disorders of the Immune system (Chapter 3)". in Stephen J. McPhee. Pathophysiology of Disease: An Introduction to Clinical Medicine (5 ed.). Lange Medical Books/McGraw-Hill. pp. 32–58. ISBN 978-0-07-110523-1.
- Nairn, Roderick (2004). "Immunology (Chapter 8)". in Geo F. Brooks. Jawetz, Melnick, & Adelberg's Medical Microbiology (Twenty-Third Edition International ed.). Lange publications/McGraw-Hill. pp. 133–135, 138–139. ISBN 978-0-07-123983-7.
External links
 |