Biology:Phagosome
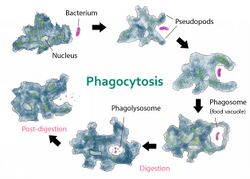
In cell biology, a phagosome is a vesicle formed around a particle engulfed by a phagocyte via phagocytosis. Professional phagocytes include macrophages, neutrophils, and dendritic cells (DCs).[1]
A phagosome is formed by the fusion of the cell membrane around a microorganism, a senescent cell or an apoptotic cell. Phagosomes have membrane-bound proteins to recruit and fuse with lysosomes to form mature phagolysosomes. The lysosomes contain hydrolytic enzymes and reactive oxygen species (ROS) which kill and digest the pathogens. Phagosomes can also form in non-professional phagocytes, but they can only engulf a smaller range of particles, and do not contain ROS. The useful materials (e.g. amino acids) from the digested particles are moved into the cytosol, and waste is removed by exocytosis. Phagosome formation is crucial for tissue homeostasis and both innate and adaptive host defense against pathogens.
However, some bacteria can exploit phagocytosis as an invasion strategy. They either reproduce inside of the phagolysosome (e.g. Coxiella spp.)[2] or escape into the cytoplasm before the phagosome fuses with the lysosome (e.g. Rickettsia spp.).[3] Many Mycobacteria, including Mycobacterium tuberculosis[4][5] and Mycobacterium avium paratuberculosis,[6] can manipulate the host macrophage to prevent lysosomes from fusing with phagosomes and creating mature phagolysosomes. Such incomplete maturation of the phagosome maintains an environment favorable to the pathogens inside it.[7]
Formation
Phagosomes are large enough to degrade whole bacteria, or apoptotic and senescent cells, which are usually >0.5μm in diameter.[8] This means a phagosome is several orders of magnitude bigger than an endosome, which is measured in nanometres.
Phagosomes are formed when pathogens or opsonins bind to a transmembrane receptor, which are randomly distributed on the phagocyte cell surface. Upon binding, "outside-in" signalling triggers actin polymerisation and pseudopodia formation, which surrounds and fuses behind the microorganism. Protein kinase C, phosphoinositide 3-kinase, and phospholipase C (PLC) are all needed for signalling and controlling particle internalisation.[9] More cell surface receptors can bind to the particle in a zipper-like mechanism as the pathogen is surrounded, increasing the binding avidity.[10] Fc receptor (FcR), complement receptors (CR), mannose receptor and dectin-1 are phagocytic receptors, which means that they can induce phagocytosis if they are expressed in non-phagocytic cells such as fibroblasts.[11] Other proteins such as Toll-like receptors are involved in pathogen pattern recognition and are often recruited to phagosomes but do not specifically trigger phagocytosis in non-phagocytic cells, so they are not considered phagocytic receptors.
Opsonisation
Opsonins are molecular tags such as antibodies and complements that attach to pathogens and up-regulate phagocytosis. Immunoglobulin G (IgG) is the major type of antibody present in the serum. It is part of the adaptive immune system, but it links to the innate response by recruiting macrophages to phagocytose pathogens. The antibody binds to microbes with the variable Fab domain, and the Fc domain binds to Fc receptors (FcR) to induce phagocytosis.
Complement-mediated internalisation has much less significant membrane protrusions, but the downstream signalling of both pathways converge to activate Rho GTPases.[12] They control actin polymerisation which is required for the phagosome to fuse with endosomes and lysosomes.
Non-phagocytic cells
Other non-professional phagocytes have some degree of phagocytic activity, such as thyroid and bladder epithelial cells that can engulf erythrocytes and retinal epithelial cells that internalise retinal rods.[8] However non-professional phagocytes do not express specific phagocytic receptors such as FcR and have a much lower rate of internalisation.
Some invasive bacteria can also induce phagocytosis in non-phagocytic cells to mediate host uptake. For example, Shigella can secrete toxins that alter the host cytoskeleton and enter the basolateral side of enterocytes.[13]
Structure
As the membrane of the phagosome is formed by the fusion of the plasma membrane, the basic composition of the phospholipid bilayer is the same. Endosomes and lysosomes then fuse with the phagosome to contribute to the membrane, especially when the engulfed particle is very big, such as a parasite.[14] They also deliver various membrane proteins to the phagosome and modify the organelle structure.
Phagosomes can engulf artificial low-density latex beads and then purified along a sucrose concentration gradient, allowing the structure and composition to be studied.[15] By purifying phagosomes at different time points, the maturation process can also be characterised. Early phagosomes are characterised by Rab5, which transition into Rab7 as the vesicle matures into late phagosomes.
Maturation process
The nascent phagosome is not inherently bactericidal. As it matures, it becomes more acidic from pH 6.5 to pH 4, and gains characteristic protein markers and hydrolytic enzymes. The different enzymes function at various optimal pH, forming a range so they each work in narrow stages of the maturation process. Enzyme activity can be fine-tuned by modifying the pH level, allowing for greater flexibility. The phagosome moves along microtubules of the cytoskeleton, fusing with endosomes and lysosomes sequentially in a dynamic "kiss-and-run" manner.[16] This intracellular transport depends on the size of the phagosomes. Larger organelles (with a diameter of about 3 μm) are transported very persistently from the cell periphery towards the perinuclear region whereas smaller organelles (with a diameter of about 1 μm) are transported more bidirectionally back and forth between cell center and cell periphery.[17] Vacuolar proton pumps (v-ATPase) are delivered to the phagosome to acidify the organelle compartment, creating a more hostile environment for pathogens and facilitating protein degradation. The bacterial proteins are denatured in low pH and become more accessible to the proteases, which are unaffected by the acidic environment. The enzymes are later recycled from the phagolysosome before egestion so they are not wasted. The composition of the phospholipid membrane also changes as the phagosome matures.[15]
Fusion may take minutes to hours depending on the contents of the phagosome; FcR or mannose receptor-mediated fusion last less than 30 minutes, but phagosomes containing latex beads may take several hours to fuse with lysosomes.[8] It is suggested that the composition of the phagosome membrane affects the rate of maturation. Mycobacterium tuberculosis has a very hydrophobic cell wall, which is hypothesised to prevent membrane recycling and recruitment of fusion factors, so the phagosome does not fuse with lysosomes and the bacterium avoids degradation.[18]
Smaller lumenal molecules are transferred by fusion faster than larger molecules, which suggests that a small aqueous channel forms between the phagosome and other vesicles during "kiss-and-run", through which only limited exchange is allowed.[8]
Fusion regulation
Shortly after internalisation, F-actin depolymerises from the newly formed phagosome so it becomes accessible to endosomes for fusion and delivery of proteins.[8] The maturation process is divided into early and late stages depending on characteristic protein markers, regulated by small Rab GTPases. Rab5 is present on early phagosomes, and controls the transition to late phagosomes marked by Rab7.[19]
Rab5 recruits PI-3 kinase and other tethering proteins such as Vps34 to the phagosome membrane, so endosomes can deliver proteins to the phagosome. Rab5 is partially involved in the transition to Rab7, via the CORVET complex and the HOPS complex in yeast.[19] The exact maturation pathway in mammals is not well understood, but it is suggested that HOPS can bind Rab7 and displace the guanosine nucleotide dissociation inhibitor (GDI).[20] Rab11 is involved in membrane recycling.[21]
Phagolysosome
The phagosome fuses with lysosomes to form a phagolysosome, which has various bactericidal properties. The phagolysosome contains reactive oxygen and nitrogen species (ROS and RNS) and hydrolytic enzymes. The compartment is also acidic due to proton pumps (v-ATPases) that transport H+ across the membrane, used to denature the bacterial proteins.
The exact properties of phagolysosomes vary depending on the type of phagocyte. Those in dendritic cells have weaker bactericidal properties than those in macrophages and neutrophils. Also, macrophages are divided into pro-inflammatory "killer" M1 and "repair" M2. The phagolysosomes of M1 can metabolise arginine into highly reactive nitric oxide, while M2 use arginine to produce ornithine to promote cell proliferation and tissue repair.[22]
Function
Pathogen degradation
Macrophages and neutrophils are professional phagocytes in charge of most of the pathogen degradation, but they have different bactericidal methods. Neutrophils have granules that fuse with the phagosome. The granules contain NADPH oxidase and myeloperoxidase, which produce toxic oxygen and chlorine derivatives to kill pathogens in an oxidative burst. Proteases and anti-microbial peptides are also released into the phagolysosome. Macrophages lack granules, and rely more on phagolysosome acidification, glycosidases, and proteases to digest microbes.[21] Phagosomes in dendritic cells are less acidic and have much weaker hydrolytic activity, due to a lower concentration of lysosomal proteases and even the presence of protease inhibitors.
Inflammation
Phagosome formation is tied to inflammation via common signalling molecules. PI-3 kinase and PLC are involved in both the internalisation mechanism and triggering inflammation.[9] The two proteins, along with Rho GTPases, are important components of the innate immune response, inducing cytokine production and activating the MAP kinase signalling cascade. Pro-inflammatory cytokines including IL-1β, IL-6, TNFα, and IL-12 are all produced.[8]
The process is tightly regulated and the inflammatory response varies depending on the particle type within the phagosome. Pathogen-infected apoptotic cells will trigger inflammation, but damaged cells that are degraded as part of the normal tissue turnover do not. The response also differs according to the opsonin-mediated phagocytosis. FcR and mannose receptor-mediated reactions produce pro-inflammatory reactive oxygen species and arachidonic acid molecules, but CR-mediated reactions do not result in those products.[8]
Antigen presentation
Immature dendritic cells (DCs) can phagocytose, but mature DCs cannot due to changes in Rho GTPases involved in cytoskeleton remodelling.[21] The phagosomes of DCs are less hydrolytic and acidic than those of macrophages and neutrophils, as DCs are mainly involved in antigen presentation rather than pathogen degradation. They need to retain protein fragments of a suitable size for specific bacterial recognition, so the peptides are only partially degraded.[21] Peptides from the bacteria are trafficked to the Major Histocompatibility Complex (MHC). The peptide antigens are presented to lymphocytes, where they bind to T-cell receptors and activates T-cells, bridging the gap between innate and adaptive immunity.[9] This is specific to mammals, birds, and jawed fish, as insects do not have adaptive immunity.[23]
Nutrient
Ancient single-celled organisms such as amoeba use phagocytosis as a way to acquire nutrients, rather than an immune strategy. They engulf other smaller microbes and digest them within the phagosome of around one bacterium per minute, which is much faster than professional phagocytes.[24] For the soil amoeba Dictyostelium discoideum, their main food source is the bacteria Legionella pneumophila, which causes Legionnaire's disease in humans.[25] Phagosome maturation in amoeba is very similar to that in macrophages, so they are used as a model organism to study the process.[16]
Tissue clearance
Phagosomes degrade senescent cells and apoptotic cells to maintain tissue homeostasis. Erythrocytes have one of the highest turnover rates in the body, and they are phagocytosed by macrophages in the liver and spleen. In the embryo, the process of removing dead cells is not well-characterised, but it is not performed by macrophages or other cells derived from hematopoietic stem cells.[26] It is only in the adult that apoptotic cells are phagocytosed by professional phagocytes. Inflammation is only triggered by certain pathogen- or damage-associated molecular patterns (PAMPs or DAMPs), the removal of senescent cells is non-inflammatory.[14]
Autophagosome
Autophagosomes are different from phagosomes in that they are mainly used to selectively degrade damaged cytosolic organelles such as mitochondria (mitophagy). However, when the cell is starved or stressed, autophagosomes can also non-selectively degrade organelles to provide the cell with amino acids and other nutrients.[27] Autophagy is not limited to professional phagocytes, it is first discovered in rat hepatocytes by cell biologist Christian de Duve.[28] Autophagosomes have a double membrane, the inner one from the engulfed organelle, and the outer membrane is speculated to be formed from the endoplasmic reticulum or the ER-Golgi Intermediate Compartment (ERGIC).[29] The autophagosome also fuses with lysosomes to degrade its contents. When M. tuberculosis inhibit phagosome acidification, Interferon gamma can induce autophagy and rescue the maturation process.[30]
Bacterial evasion and manipulation
Many bacteria have evolved to evade the bactericidal properties of phagosomes or even exploit phagocytosis as an invasion strategy.
- Mycobacterium tuberculosis target M2 macrophages at the lower parts of the respiratory pathway, which do not produce ROS.[31] M. tuberculosis can also manipulate the signalling pathways by secreting phosphatases such as PtpA and SapM, which disrupt protein recruitment and block phagosome acidification.[8][32]
- Legionella pneumophila can re-model the phagosome membrane to imitate vesicles in other parts of the secretory pathway, so lysosomes do not recognise the phagosome and do not fuse with it. The bacterium secretes toxins that interfere with host trafficking, so the Legionella-containing vacuole recruits membrane proteins usually found on the endoplasmic reticulum or the ERGIC.[33] This re-directs secretory vesicles to the modified phagosome and deliver nutrients to the bacterium.
- Listeria monocytogenes secretes a pore-forming protein listeriolysin O so the bacterium can escape the phagosome into the cytosol. Listeriolysin is activated by the acidic environment of the phagosome.[34] In addition, Listeria secrete two phospholipase C enzymes that facilitate in phagosome escape.
See also
References
- ↑ Robinson & Babcock 1998, p. 187 and Ernst & Stendahl 2006, pp. 7–10
- ↑ "Biochemical stratagem for obligate parasitism of eukaryotic cells by Coxiella burnetii". Proceedings of the National Academy of Sciences of the United States of America 78 (5): 3240–4. May 1981. doi:10.1073/pnas.78.5.3240. PMID 6942430.
- ↑ "Rickettsia Species (As Organisms)". Annual Review of Microbiology 44: 131–153. 1990. doi:10.1146/annurev.micro.44.1.131. PMID 2252380.
- ↑ "Immune control of tuberculosis by IFN-gamma-inducible LRG-47". Science 302 (5645): 654–9. October 2003. doi:10.1126/science.1088063. PMID 14576437. Bibcode: 2003Sci...302..654M.
- ↑ "A membrane protein preserves intrabacterial pH in intraphagosomal Mycobacterium tuberculosis". Nature Medicine 14 (8): 849–54. August 2008. doi:10.1038/nm.1795. PMID 18641659.
- ↑ "Characterization of the intracellular survival of Mycobacterium avium ssp. paratuberculosis: phagosomal pH and fusogenicity in J774 macrophages compared with other mycobacteria". Cellular Microbiology 3 (8): 551–66. August 2001. doi:10.1046/j.1462-5822.2001.00139.x. PMID 11488816.
- ↑ "How does Mycobacterium avium subsp. paratuberculosis resist intracellular degradation?". The Veterinary Quarterly 23 (4): 153–62. November 2001. doi:10.1080/01652176.2001.9695105. PMID 11765232.
- ↑ 8.0 8.1 8.2 8.3 8.4 8.5 8.6 8.7 "Mechanisms of phagocytosis in macrophages". Annual Review of Immunology 17 (1): 593–623. April 1999. doi:10.1146/annurev.immunol.17.1.593. PMID 10358769.
- ↑ 9.0 9.1 9.2 "Phagocytosis and the inflammatory response". The Journal of Infectious Diseases 187 Suppl 2 (s2): S340-5. June 2003. doi:10.1086/374747. PMID 12792849.
- ↑ "Integrin-dependent phagocytosis: spreading from microadhesion to new concepts". Journal of Cell Science 121 (11): 1773–83. June 2008. doi:10.1242/jcs.018036. PMID 18492791.
- ↑ "Phagocytosis of microbes: complexity in action". Annual Review of Immunology 20 (1): 825–52. April 2002. doi:10.1146/annurev.immunol.20.103001.114744. PMID 11861619.
- ↑ "Differences in the mode of phagocytosis with Fc and C3 receptors in macrophages". Scandinavian Journal of Immunology 6 (8): 797–807. August 1977. doi:10.1111/j.1365-3083.1977.tb02153.x. PMID 561436.
- ↑ "Shigella flexneri Interactions with the Basolateral Membrane Domain of Polarized Model Intestinal Epithelium: Role of Lipopolysaccharide in Cell Invasion and in Activation of the Mitogen-Activated Protein Kinase ERK". Infection and Immunity 70 (3): 1150–8. March 2002. doi:10.1128/IAI.70.3.1150-1158.2002. PMID 11854195.
- ↑ 14.0 14.1 "Phagocytosis: the convoluted way from nutrition to adaptive immunity". Immunological Reviews 207 (1): 158–65. October 2005. doi:10.1111/j.0105-2896.2005.00319.x. PMID 16181334.
- ↑ 15.0 15.1 "Molecular characterization of phagosomes". The Journal of Biological Chemistry 269 (51): 32194–200. December 1994. doi:10.1016/S0021-9258(18)31620-X. PMID 7798218.
- ↑ 16.0 16.1 "High-resolution dissection of phagosome maturation reveals distinct membrane trafficking phases". Molecular Biology of the Cell 13 (10): 3508–20. October 2002. doi:10.1091/mbc.E02-04-0206. PMID 12388753.
- ↑ "Phagosomal transport depends strongly on phagosome size". Scientific Reports 7 (1): 17068. December 2017. doi:10.1038/s41598-017-17183-7. PMID 29213131. Bibcode: 2017NatSR...717068K.
- ↑ "Phagosome maturation and fusion with lysosomes in relation to surface property and size of the phagocytic particle". European Journal of Cell Biology 74 (1): 49–62. September 1997. PMID 9309390.
- ↑ 19.0 19.1 "How nascent phagosomes mature to become phagolysosomes". Trends in Immunology 33 (8): 397–405. August 2012. doi:10.1016/j.it.2012.03.003. PMID 22560866.
- ↑ "Identification of two evolutionarily conserved genes regulating processing of engulfed apoptotic cells". Nature 464 (7289): 778–82. April 2010. doi:10.1038/nature08853. PMID 20305638. Bibcode: 2010Natur.464..778K.
- ↑ 21.0 21.1 21.2 21.3 "Phagocytosis and antigen presentation in dendritic cells". Immunological Reviews 219 (1): 143–56. October 2007. doi:10.1111/j.1600-065X.2007.00552.x. PMID 17850487.
- ↑ "Anatomy of a discovery: m1 and m2 macrophages". Frontiers in Immunology 6: 212. 5 May 2015. doi:10.3389/fimmu.2015.00212. PMID 25999950.
- ↑ "The adaptive immune system of fish". Developments in Biological Standardization 90: 15–21. 1997. PMID 9270830.
- ↑ "Eat, kill or die: when amoeba meets bacteria". Current Opinion in Microbiology 11 (3): 271–6. June 2008. doi:10.1016/j.mib.2008.05.005. PMID 18550419.
- ↑ "Intracellular growth of Legionella pneumophila in Dictyostelium discoideum, a system for genetic analysis of host-pathogen interactions". Infection and Immunity 68 (5): 2939–47. May 2000. doi:10.1128/iai.68.5.2939-2947.2000. PMID 10768992.
- ↑ "Origins and functions of phagocytes in the embryo". Experimental Hematology 28 (6): 601–11. June 2000. doi:10.1016/s0301-472x(00)00157-0. PMID 10880746. https://www.pure.ed.ac.uk/ws/files/12628290/Origins_and_functions_of_phagocytes_in_the_embryo.pdf.
- ↑ "Mitophagy: mechanisms, pathophysiological roles, and analysis". Biological Chemistry 393 (7): 547–64. July 2012. doi:10.1515/hsz-2012-0119. PMID 22944659.
- ↑ Castro-Obregon, Susana (2010). "The Discovery of Lysosomes and Autophagy". Nature Education 3 (9): 49. http://www.nature.com/scitable/topicpage/the-discovery-of-lysosomes-and-autophagy-14199828.
- ↑ "The ER-Golgi intermediate compartment feeds the phagophore membrane". Autophagy 10 (1): 170–2. January 2014. doi:10.4161/auto.26787. PMID 24220263.
- ↑ "Autophagy is a defense mechanism inhibiting BCG and Mycobacterium tuberculosis survival in infected macrophages". Cell 119 (6): 753–66. December 2004. doi:10.1016/j.cell.2004.11.038. PMID 15607973.
- ↑ "Mycobacteria manipulate macrophage recruitment through coordinated use of membrane lipids". Nature 505 (7482): 218–22. January 2014. doi:10.1038/nature12799. PMID 24336213.
- ↑ "Mycobacterium tuberculosis-secreted phosphatases: from pathogenesis to targets for TB drug development". Trends in Microbiology 21 (2): 100–9. February 2013. doi:10.1016/j.tim.2012.09.002. PMID 23084287.
- ↑ Roy, Craig R.; Kagan, Jonathan C. (1 January 2013) (in en). Evasion of Phagosome Lysosome Fusion and Establishment of a Replicative Organelle by the Intracellular Pathogen Legionella pneumophila. Landes Bioscience. https://www.ncbi.nlm.nih.gov/books/NBK6111/.
- ↑ "The cell biology of Listeria monocytogenes infection: the intersection of bacterial pathogenesis and cell-mediated immunity". The Journal of Cell Biology 158 (3): 409–14. August 2002. doi:10.1083/jcb.200205009. PMID 12163465.
 |