Earth:Sea ice microbial communities
Sea Ice Microbial Communities (SIMCO) refer to groups of microorganisms living within and at the interfaces of sea ice at the poles. The ice matrix they inhabit has strong vertical gradients of salinity, light, temperature and nutrients. Sea ice chemistry is most influenced by the salinity of the brine which affects the pH and the concentration of dissolved nutrients and gases. The brine formed during the melting sea ice creates pores and channels in the sea ice in which these microbes can live. As a result of these gradients and dynamic conditions, a higher abundance of microbes are found in the lower layer of the ice, although some are found in the middle and upper layers. Despite this extreme variability in environmental conditions, the taxonomical community composition tends to remain consistent throughout the year, until the ice melts.[1]
Much of the knowledge concerning the community diversity of the sea ice is known through genetic analyses and next-generation sequencing. In both the Arctic and Antarctic, Alphaproteobacteria, Gammaproteobacteria and Flavobacteriia are the common bacterial classes found. Most sea ice Archaea belong to the phylum Nitrososphaerota while most of the protists belong to one of 3 supergroups: Alveolata, Stramenopile and Rhizaria. The abundance of living cells within and on sea ice ranges from 104-108 cells/mL.[1] These microbial communities play a significant role in the microbial loop as well as in global biogeochemical cycles. Sea ice communities are important because they provide an energy source for higher trophic levels, they contribute to primary production and they provide a net influx of Carbon in the oceans at the poles.[2]
Habitat
Sea ice matrix: chemical and physical properties
Sea ice formation and physical properties
The autumnal decrease in atmospheric temperatures in the Arctic and Antarctic leads to the formation of a surface layer of ice crystals called frazil ice. A mixture of salts, nutrients and dissolved organic matter (DOM) known as brine is expelled when frazil ice solidifies to form sea ice. Brine permeates through the ice cover and creates a network of channels and pores. This process forms an initial semisolid matrix of approximately 1 meter in thickness with strong temperature, salinity, light and nutrient gradients.[3]
Since thickening of the sea ice during winter months results in more salts being expelled from the frazil ice, atmospheric temperatures are strongly and negatively correlated to brine salinity. The sea ice-seawater interface temperature is maintained at the freezing point of seawater (~1.8 °C) while the sea ice-air interface reflects more the current atmospheric temperature.[4] Brine salinity can increase to as much as 100 PSU when sea ice temperature reaches ~3 °C below the freezing point of seawater.[5] Brine temperature typically ranges from -1.9 to -6.7 °C in the winter.[6] Sea ice temperatures fluctuate in response to irradiance and atmospheric temperatures, but also change in response to the volume of snowfall. Accumulating snow on the ice cover combined with harsh atmospheric conditions can lead to the formation of a snowpack layer that absorbs UV radiations and provides insulation to the bottom ice layer. The fraction of irradiance reaching the sea ice matrix is thus also controlled by the amount of snowfall and varies from <0.01% to 5% depending on the thickness and density of the snowpack.[4]
The surface of sea ice also allows the formation of frost flowers, which have their own unique microbial communities.[1]
Carbon species, nutrients and gases
The fluctuation of brine salinity, which is controlled by atmospheric temperatures, is the single-most influential factor on the chemistry of the sea ice matrix. The solubility of carbon dioxide and oxygen, two biologically essential gases, decreases in higher salinity solutions. This can result in hypoxia within high heterotrophic activity regions of the sea ice matrix. Regions of high photosynthetic activity often exhibit internal depletion of inorganic carbon compound and hyperoxia. These conditions have the potential to elevate brine pH and to further contribute to the creation of an extreme environment. In these conditions, high concentrations of DOM and ammonia and low concentrations of nutrients often characterize the ice matrix.[7]
High brine salinity combined with an elevated pH reduces the rate at which gases and inorganic nutrients diffuse into the ice matrix.[6] The concentration of nutrients such as nitrate, phosphate and silicate inside the sea ice matrix relies largely on the diffusive influx from the sea ice-water interface and to some extent on the atmospheric deposits on the sea ice-air interface.[5] Iron concentrations in the Southern Ocean ice cover are thought to be regulated by the amount of new iron supply at the time of ice formation and were shown to be reduced during late winter.[8]
The chemical properties of the sea ice matrix are highly complex and depend on the interaction between the internal sea ice biological assemblage as well as external physical factors.[4] Winters are typically characterized by moderate oxygen levels that are accompanied by nutrient and inorganic carbon concentrations that are not growth limiting to phytoplankton. Summers are typically characterized by high oxygen levels that are accompanied by a depletion of nutrients and inorganic carbon. Because of its diffusive interaction with seawater, the lower part of the sea ice matrix is typically characterized by higher nutrient concentrations.[6]
Colonization
Microorganisms present in the surface seawater during fall are integrated in the brine solution during ice formation. A small proportion of the initial microbial population colonizes the ice matrix while the rest is expelled with brine.[5] Studies have shown that sea ice microbial retention can be enhanced by the presence of extracellular polymeric substance/polysaccharides (EPS) on the walls of the brine channels. EPS are proteins expressed on the cell walls of microorganism such as algae. They improve the cell adherence to surfaces and when found in sufficient concentration, are thought to play a role in recruiting other organisms such as microbes.[5]
Airborne microorganisms make up a significant proportion of the microbial input to the ice matrix. Microorganisms located in the sea or in the ice matrix brine can be incorporated in falling snow or in aerosols, and subsequently transported by strong winds such as the West Wind Drift that causes the Antarctic Circumpolar Current. These airborne microorganisms can originate from terrestrial environment and marine environment, thus contributing to the diversity of the SIMCO.[5]
Distribution
Spatial Distribution
Microbes colonizing the Antarctic sea ice are eventually incorporated in the pore spaces and brine channels of the ice matrix, but can also inhabit the ice-seawater interface.[9] Pore spaces in the matrix lose their ability to exchange nutrients, DOM and microorganisms with brine at approximately -5 °C. This suggests that the Antarctic microbial community is fluid along the ice matrix during fall and spring and that it is restricted during winter.[9]
In the Arctic, brine channels are also inhabited by bacteria. Channels as small as ≤200 μm offer a spatial refuge with microbial community concentrations of 1-2 orders of magnitude higher than in the remaining channel network.[10]
Both the Antarctic and Arctic sea ice environments present strong vertical gradients of salinity, temperature, light, nutrients and DOM. These gradients were shown to induce strong vertical stratification in bacterial communities throughout the ice layer.[11][12] Microbial abundance declines significantly with depth in the upper and middle ice, but not in the lowest, suggesting that much of the prokaryotic bacterial community is resistant to extreme environmental conditions.[11] Heterotrophic bacteria were also shown to be more abundant at the bottom of the ice layer in zones of greater algae concentration, which characterized by higher DOM and nutrient concentrations.[12]
Temporal Distribution
The temporal distribution of microbial community composition in the Antarctic and Arctic sea ice does not present significant seasonal variability, despite extremes in environmental conditions. Previous studies of sea ice habitats have shown that the composition of SIMCO in early fall is identical to the source seawater community.[13] The microbial community composition does not seem to change significantly in fall and winter, despite the extreme variability in irradiance, temperature, salinity and nutrient concentrations. In contrast, the abundance within the SIMCO is reduced throughout the winter as resources become limiting. Studies have shown that sea ice microalgae provide a platform and organic nutrient source for bacterial growth, therefore increasing community diversity and abundance.[13][14] It has also been proven that microbes produce extracellular polymeric substances (EPS) to help retain nutrients and survive under high salinity and low temperature conditions.[5]
The increase in irradiance levels in late spring promotes ice algal photosynthesis which in turn affects the microbial community abundance and composition. While most of the sea ice cover melts in late spring in the Antarctic and Arctic, multiyear sea ice occasionally persists when late spring and summer temperatures are lower than average. This suggests that certain microbial lineages may have adapted more efficiently to the extreme conditions of sea ice environments. Temporal abundance can also be affected by the thickness of the annual ice cover and seasonal temperature variations. The ice cover thickness was shown to regulate microbial production and the temperature of the ice matrix through layer insulation.[15]
Community composition
A majority of the information on sea ice microbial community composition comes from 16S ribosomal RNA taxonomic marker genes and metagenomic analyses.[9] Next-generation sequencing has allowed researcher to identify and quantify microbial communities, and to gain a more complete understanding of its structure.
Bacteria
Arctic
Metagenomic studies of Arctic sea ice show classes Alphaproteobacteria, Gammaproteobacteria and Flavobacteria. Within the Flavobacteriia class the genera Polaribacter, Psychrobacter, Psychroflexus, and Flavobacterium are the most common. Within Gammaproteobacteria the genera Glaciecola and Colwellia are the most common.[1] Also found in Arctic sea ice samples were bacteria of the following classes and phyla: "Opitutae", Bacilli, "Cyanobacteria", Betaproteobacteria, Sphingobacteria, and Aquificota.[1]
Antarctic
Metagenomic studies of the Ross Sea illustrate the high abundance of aerobic anoxygenic phototrophic bacteria in sea ice environments.[16] These specialists were shown to mostly belong to the Alphaproteobacteria class. Genera of the Alphaproteobacteria class were shown to include Loktanella, Octadecabacter, Roseobacter, Sulfitobacter and Methylobacterium and to agree with previous phylogenetic analyses of sea ice around the Antarctic. A study of the SIMCO 16S ribosomal RNA at Cape Hallett in the Antarctic has shown that aerobic oxygenic phototrophic bacteria may be equally abundant.[17]
Members of the Gammaproteobacteria and Flaviobacteria classes were also shown to be abundant within the ice matrix, and thus to be adapted to the sea ice conditions.[18] Genera of the Gammaproteobacteria class found in the Ross Sea and around the Antarctic waters include Colwellia, Marionomonas, Pseudoalteromonas and Psychrobacter.[17][19] The orders Chlamydiales and Verrucomicrobiales were also found in the sea ice microbial assemblage of these locations. The predominance of Gammaproteobacteria in sea ice around the globe have been reported by many studies. A large proportion of the identified SIMCO in these studies were shown to belong to phylotypes associated with heterotrophic taxa[20]
While this gives researchers an insight into the microbial community composition of the Antarctic sea ice, there are clear shifts between locations in the Southern Ocean. These shifts are attributed to biological and physical forcing factors. These factors include the composition of the microbial communities in place at the moment of sea ice formation, and the regional weather and wind patterns affecting the transport of snow and aerosols.[21]
Archaea
Studies of the 16s ribosomal RNA subunits found in the sea ice cover of Terra Nova Bay have shown that archaea consist of ≤ 6.6% of the total prokaryotic community in this environment. 90.8% of this archaeal community belonged to the phylum Nitrososphaerota (formerly Thaumarchaeota), a close parent to marine ammonia-oxidizing bacteria, while Euryarchaeota made up the rest of the community.[17] Other molecular phylogenetic analyses of the SIMCO have detected no trace of the archaeal domain.[20]
Protists
Metagenomic studies of Arctic sea ice using 454 sequencing of 18S rDNA and 18S rRNA. These studies showed dominance of three supergroups: Alveolata, Stramenophile, and Rhizaria. Within the Alveolates most common were Ciliates, and Dinoflagellates. Within the Stramenophile group most organisms were classified as Bacillariophyceae. Finally most of the Rhizarians were classified as from Thecofilosea.[2]
Adaptation
Studies have shown that high concentrations of microbial cryoprotective exopolymer (EPS) were found in the sea ice brine. These EPS were shown to correlate with a stable microbial community composition throughout the winter season.[9] They are thought to play an important role in sea ice environments where they act as a buffer and cryoprotectant against high salinity and ice-crystal damage. These exopolymers are believed to constitute a microbial adaptation to low temperatures in extreme environments.[22]
Sea ice microbes have also developed anti-freeze proteins, which prevent the formation of ice crystals that could damage bacterial membranes.[1] It is common for these proteins to be rich in beta-sheets as they prevent the formation of ice crystals/[23]
Metabolic diversity
Role in the Microbial Loop
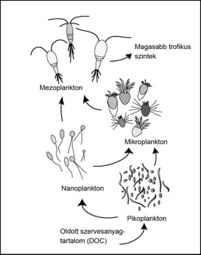
Bacteria in all environments contribute to the microbial loop, but the roles of sea ice microbial communities in the microbial loop differ due to the rapidly changing environmental conditions found in the Arctic and Antarctic. Sea ice algae contribute 10%–28% of the total primary production in ice-covered regions of the Antarctic.[24] Microalgae provide a vital source of nutrition for juvenile zooplankton such as the Antarctic krill Euphausia superba in the winter.[24] DOM derived from phototrophic microalgae is crucial to the microbial loop, by serving as a growth substrate for heterotrophic bacteria.[24]
The microbial loop functions differently in sea ice, as compared to oligotrophic or temperate waters. Animals found in the extreme polar environments depend on the high bacterial production as a food source, despite the slow turnover of DOM.[25] The microbial production of ammonium in nitrate-rich Antarctic waters may provide the necessary reductants for nitrogen fixation, increasing primary productivity of light-limited phytoplankton.[25] Phytoflagellates and diatoms found in the Antarctic pelagic environment are directly digestible by metozoan herbivores.[25]
See also
References
- ↑ 1.0 1.1 1.2 1.3 1.4 1.5 Boetius, Antje; Anesio, Alexandre M.; Deming, Jody W.; Mikucki, Jill A.; Rapp, Josephine Z. (November 1, 2015). "Microbial ecology of the cryosphere: sea ice and glacial habitats". Nature Reviews Microbiology 13 (11): 677–690. doi:10.1038/nrmicro3522. PMID 26344407.
- ↑ 2.0 2.1 Stecher, Aniqu; Neuhaus, Stefan; Lange, Benjamin; Frickenhaus, Stephan; Beszteri, Bank; Kroth, Peter G.; Valentin, Klaus (October 22, 2015). "rRNA and rDNA based assessment of sea ice protest biodiversity from the central Arctic Ocean". European Journal of Phycology 51 (1): 31–46. doi:10.1080/09670262.2015.1077395. http://epic.awi.de/38995/1/Stecher_et_al_2015.pdf.
- ↑ Eicken, Hajo (1992-10-15). "Salinity profiles of Antarctic sea ice: Field data and model results". Journal of Geophysical Research: Oceans 97 (C10): 15545–15557. doi:10.1029/92jc01588. Bibcode: 1992JGR....9715545E.
- ↑ 4.0 4.1 4.2 Thomas, D. N.; Dieckmann, G. S. (2002-01-25). "Antarctic Sea ice--a habitat for extremophiles". Science 295 (5555): 641–644. doi:10.1126/science.1063391. PMID 11809961. Bibcode: 2002Sci...295..641T. http://epic.awi.de/5290/1/Tho2002b.pdf.
- ↑ 5.0 5.1 5.2 5.3 5.4 5.5 Ewert, Marcela; Deming, Jody W. (2013-03-28). "Sea Ice Microorganisms: Environmental Constraints and Extracellular Responses". Biology 2 (2): 603–628. doi:10.3390/biology2020603. PMID 24832800.
- ↑ 6.0 6.1 6.2 Gleitz, Markus; Loeff, Michiel Rutgers v.d.; Thomas, David N.; Dieckmann, Gerhard S.; Millero, Frank J. (1995). "Comparison of summer and winter inorganic carbon, oxygen and nutrient concentrations in Antarctic sea ice brine". Marine Chemistry 51 (2): 81–91. doi:10.1016/0304-4203(95)00053-t.
- ↑ Rysgaard, S.; Glud, R. N.; Sejr, M. K.; Bendtsen, J.; Christensen, P. B. (2007-03-01). "Inorganic carbon transport during sea ice growth and decay: A carbon pump in polar seas". Journal of Geophysical Research: Oceans 112 (C3): C03016. doi:10.1029/2006jc003572. Bibcode: 2007JGRC..112.3016R.
- ↑ Lannuzel, Delphine; Schoemann, Véronique; de Jong, Jeroen; Pasquer, Bénédicte; van der Merwe, Pier; Masson, Florence; Tison, Jean-Louis; Bowie, Andrew (2010-09-01). "Distribution of dissolved iron in Antarctic sea ice: Spatial, seasonal, and inter-annual variability". Journal of Geophysical Research: Biogeosciences 115 (G3): G03022. doi:10.1029/2009jg001031. Bibcode: 2010JGRG..115.3022L.
- ↑ 9.0 9.1 9.2 9.3 Bowman, Jeff S. (13 October 2015). "The relationship between sea ice bacterial community structure and biogeochemistry: A synthesis of current knowledge and known unknowns". Elementa: Science of the Anthropocene 3: 000072. doi:10.12952/journal.elementa.000072.
- ↑ Krembs, C; Gradinger, R; Spindler, M (1 January 2000). "Implications of brine channel geometry and surface area for the interaction of sympagic organisms in Arctic sea ice". Journal of Experimental Marine Biology and Ecology 243 (1): 55–80. doi:10.1016/S0022-0981(99)00111-2.
- ↑ 11.0 11.1 Collins, R. Eric; Carpenter, Shelly D; Deming, Jody W (1 December 2008). "Spatial heterogeneity and temporal dynamics of particles, bacteria, and pEPS in Arctic winter sea ice". Journal of Marine Systems 74 (3–4): 902–917. doi:10.1016/j.jmarsys.2007.09.005. Bibcode: 2008JMS....74..902C.
- ↑ 12.0 12.1 Cowie, ROM; Williams, GJ; Maas, EW; Voyles, KM; Ryan, KG (2 October 2014). "Antarctic sea-ice microbial communities show distinct patterns of zonation in response to algal-derived substrates". Aquatic Microbial Ecology 73 (2): 123–134. doi:10.3354/ame01710.
- ↑ 13.0 13.1 Collins, R. Eric; Rocap, Gabrielle; Deming, Jody W. (25 February 2010). "Persistence of bacterial and archaeal communities in sea ice through an Arctic winter". Environmental Microbiology 12 (7): 1828–1841. doi:10.1111/j.1462-2920.2010.02179.x. PMID 20192970.
- ↑ McGrath Grossi, Sarah; Kottmeier, Steven T.; Sullivan, C. W. (September 1984). "Sea ice microbial communities. III. Seasonal abundance of microalgae and associated bacteria, Mcmurdo Sound, Antarctica". Microbial Ecology 10 (3): 231–242. doi:10.1007/bf02010937. PMID 24221145.
- ↑ Eronen-Rasimus, Eeva; Luhtanen, Anne-Mari; Rintala, Janne-Markus; Delille, Bruno; Dieckmann, Gerhard; Karkman, Antti; Tison, Jean-Louis (October 2017). "An active bacterial community linked to high chl-a concentrations in Antarctic winter-pack ice and evidence for the development of an anaerobic sea-ice bacterial community". The ISME Journal 11 (10): 2345–2355. doi:10.1038/ismej.2017.96. PMID 28708127.
- ↑ Koh, Eileen Y.; Phua, William; Ryan, Ken G. (2011-12-01). "Aerobic anoxygenic phototrophic bacteria in Antarctic sea ice and seawater". Environmental Microbiology Reports 3 (6): 710–716. doi:10.1111/j.1758-2229.2011.00286.x. PMID 23761361.
- ↑ 17.0 17.1 17.2 Cowie, Rebecca O. M.; Maas, Elizabeth W.; Ryan, Ken G. (November 22, 2011). "Archaeal diversity revealed in Antarctic sea ice". Antarctic Science 23 (6): 531–536. doi:10.1017/s0954102011000368. Bibcode: 2011AntSc..23..531C.
- ↑ Koh, Eileen Y.; Atamna-Ismaeel, Nof; Martin, Andrew; Cowie, Rebecca O. M.; Beja, Oded; Davy, Simon K.; Maas, Elizabeth W.; Ryan, Ken G. (September 2010). "Proteorhodopsin-bearing bacteria in Antarctic sea ice". Applied and Environmental Microbiology 76 (17): 5918–5925. doi:10.1128/AEM.00562-10. PMID 20601510. Bibcode: 2010ApEnM..76.5918K.
- ↑ Brinkmeyer, Robin; Knittel, Katrin; Jürgens, Jutta; Weyland, Horst; Amann, Rudolf; Helmke, Elisabeth (November 2003). "Diversity and structure of bacterial communities in Arctic versus Antarctic pack ice". Applied and Environmental Microbiology 69 (11): 6610–6619. doi:10.1128/AEM.69.11.6610-6619.2003. PMID 14602620. Bibcode: 2003ApEnM..69.6610B.
- ↑ 20.0 20.1 Brown, M. V.; Bowman, J. P. (May 2001). "A molecular phylogenetic survey of sea-ice microbial communities (SIMCO)". FEMS Microbiology Ecology 35 (3): 267–275. doi:10.1111/j.1574-6941.2001.tb00812.x. PMID 11311437.
- ↑ ARRIGO, Kevin R.; Thomas, David N. (December 2004). "Large scale importance of sea ice biology in the Southern Ocean". Antarctic Science 16 (4): 471–486. doi:10.1017/s0954102004002263. Bibcode: 2004AntSc..16..471A.
- ↑ Hodgson, E. K.; Fridovich, I. (1975-12-02). "The interaction of bovine erythrocyte superoxide dismutase with hydrogen peroxide: inactivation of the enzyme". Biochemistry 14 (24): 5294–5299. doi:10.1021/bi00695a010. PMID 49.
- ↑ Muñoz, Patricio A.; Márquez, Sebastián L.; González-Nilo, Fernando D.; Márquez-Miranda, Valeria; Blamey, Jenny M. (2017-08-07). "Structure and application of antifreeze proteins from Antarctic bacteria". Microbial Cell Factories 16 (1): 138. doi:10.1186/s12934-017-0737-2. PMID 28784139.
- ↑ 24.0 24.1 24.2 Koh, Eileen Y.; Martin, Andrew R.; McMinn, Andrew; Ryan, Ken G. (22 October 2012). "Recent Advances and Future Perspectives in Microbial Phototrophy in Antarctic Sea Ice". Biology 1 (3): 542–556. doi:10.3390/biology1030542. PMID 24832507.
- ↑ 25.0 25.1 25.2 Azam, Farooq; Smith, David C.; Hollibaugh, James T. (16 December 2016). "The role of the microbial loop in Antarctic pelagic ecosystems". Polar Research 10 (1): 239–244. doi:10.3402/polar.v10i1.6742.