Network science
| Network science | ||||
|---|---|---|---|---|
| Network types | ||||
| Graphs | ||||
|
||||
| Models | ||||
|
||||
| ||||
| Complex systems |
|---|
| Topics |
Network science is an academic field which studies complex networks such as telecommunication networks, computer networks, biological networks, cognitive and semantic networks, and social networks, considering distinct elements or actors represented by nodes (or vertices) and the connections between the elements or actors as links (or edges). The field draws on theories and methods including graph theory from mathematics, statistical mechanics from physics, data mining and information visualization from computer science, inferential modeling from statistics, and social structure from sociology. The United States National Research Council defines network science as "the study of network representations of physical, biological, and social phenomena leading to predictive models of these phenomena."[1]
Background and history
The study of networks has emerged in diverse disciplines as a means of analyzing complex relational data. The earliest known paper in this field is the famous Seven Bridges of Königsberg written by Leonhard Euler in 1736. Euler's mathematical description of vertices and edges was the foundation of graph theory, a branch of mathematics that studies the properties of pairwise relations in a network structure. The field of graph theory continued to develop and found applications in chemistry (Sylvester, 1878).
Dénes Kőnig, a Hungarian mathematician and professor, wrote the first book in Graph Theory, entitled "Theory of finite and infinite graphs", in 1936.[2]
In the 1930s Jacob Moreno, a psychologist in the Gestalt tradition, arrived in the United States. He developed the sociogram and presented it to the public in April 1933 at a convention of medical scholars. Moreno claimed that "before the advent of sociometry no one knew what the interpersonal structure of a group 'precisely' looked like" (Moreno, 1953). The sociogram was a representation of the social structure of a group of elementary school students. The boys were friends of boys and the girls were friends of girls with the exception of one boy who said he liked a single girl. The feeling was not reciprocated. This network representation of social structure was found so intriguing that it was printed in The New York Times (April 3, 1933, page 17). The sociogram has found many applications and has grown into the field of social network analysis.
Probabilistic theory in network science developed as an offshoot of graph theory with Paul Erdős and Alfréd Rényi's eight famous papers on random graphs. For social networks the exponential random graph model or p* is a notational framework used to represent the probability space of a tie occurring in a social network. An alternate approach to network probability structures is the network probability matrix, which models the probability of edges occurring in a network, based on the historic presence or absence of the edge in a sample of networks.
In 1998, David Krackhardt and Kathleen Carley introduced the idea of a meta-network with the PCANS Model. They suggest that "all organizations are structured along these three domains, Individuals, Tasks, and Resources". Their paper introduced the concept that networks occur across multiple domains and that they are interrelated. This field has grown into another sub-discipline of network science called dynamic network analysis.
More recently other network science efforts have focused on mathematically describing different network topologies. Duncan Watts and Steven Strogatz reconciled empirical data on networks with mathematical representation, describing the small-world network. Albert-László Barabási and Reka Albert discovered scale-free networks, a property that captures the fact that in real network hubs coexist with many small degree vertices, and offered a dynamical model to explain the origin of this scale-free state.
Department of Defense initiatives
The U.S. military first became interested in network-centric warfare as an operational concept based on network science in 1996. John A. Parmentola, the U.S. Army Director for Research and Laboratory Management, proposed to the Army's Board on Science and Technology (BAST) on December 1, 2003 that Network Science become a new Army research area. The BAST, the Division on Engineering and Physical Sciences for the National Research Council (NRC) of the National Academies, serves as a convening authority for the discussion of science and technology issues of importance to the Army and oversees independent Army-related studies conducted by the National Academies. The BAST conducted a study to find out whether identifying and funding a new field of investigation in basic research, Network Science, could help close the gap between what is needed to realize Network-Centric Operations and the current primitive state of fundamental knowledge of networks.
As a result, the BAST issued the NRC study in 2005 titled Network Science (referenced above) that defined a new field of basic research in Network Science for the Army. Based on the findings and recommendations of that study and the subsequent 2007 NRC report titled Strategy for an Army Center for Network Science, Technology, and Experimentation, Army basic research resources were redirected to initiate a new basic research program in Network Science. To build a new theoretical foundation for complex networks, some of the key Network Science research efforts now ongoing in Army laboratories address:
- Mathematical models of network behavior to predict performance with network size, complexity, and environment
- Optimized human performance required for network-enabled warfare
- Networking within ecosystems and at the molecular level in cells.
As initiated in 2004 by Frederick I. Moxley with support he solicited from David S. Alberts, the Department of Defense helped to establish the first Network Science Center in conjunction with the U.S. Army at the United States Military Academy (USMA). Under the tutelage of Dr. Moxley and the faculty of the USMA, the first interdisciplinary undergraduate courses in Network Science were taught to cadets at West Point.[3][4][5] In order to better instill the tenets of network science among its cadre of future leaders, the USMA has also instituted a five-course undergraduate minor in Network Science. [6]
In 2006, the U.S. Army and the United Kingdom (UK) formed the Network and Information Science International Technology Alliance, a collaborative partnership among the Army Research Laboratory, UK Ministry of Defense and a consortium of industries and universities in the U.S. and UK. The goal of the alliance is to perform basic research in support of Network- Centric Operations across the needs of both nations.
In 2009, the U.S. Army formed the Network Science CTA, a collaborative research alliance among the Army Research Laboratory, CERDEC, and a consortium of about 30 industrial R&D labs and universities in the U.S. The goal of the alliance is to develop a deep understanding of the underlying commonalities among intertwined social/cognitive, information, and communications networks, and as a result improve our ability to analyze, predict, design, and influence complex systems interweaving many kinds of networks.
Subsequently, as a result of these efforts, the U.S. Department of Defense has sponsored numerous research projects that support Network Science.
Network Classification
Deterministic Network
The definition of deterministic network is defined compared with the definition of probabilistic network. In un-weighted deterministic networks, edges either exist or not, usually we use 0 to represent non-existence of an edge while 1 to represent existence of an edge. In weighted deterministic networks, the edge value represents the weight of each edge, for example, the strength level.
Probabilistic Network
In probabilistic networks, values behind each edge represent the likelihood of the existence of each edge. For example, if one edge has a value equals to 0.9, we say the existence probability of this edge is 0.9.[7]
Network properties
Often, networks have certain attributes that can be calculated to analyze the properties & characteristics of the network. The behavior of these network properties often define network models and can be used to analyze how certain models contrast to each other. Many of the definitions for other terms used in network science can be found in Glossary of graph theory.
Size
The size of a network can refer to the number of nodes [math]\displaystyle{ N }[/math] or, less commonly, the number of edges [math]\displaystyle{ E }[/math] which (for connected graphs with no multi-edges) can range from [math]\displaystyle{ N-1 }[/math] (a tree) to [math]\displaystyle{ E_{\max} }[/math] (a complete graph). In the case of a simple graph (a network in which at most one (undirected) edge exists between each pair of vertices, and in which no vertices connect to themselves), we have [math]\displaystyle{ E_{\max}=\tbinom N2=N(N-1)/2 }[/math]; for directed graphs (with no self-connected nodes), [math]\displaystyle{ E_{\max}=N(N-1) }[/math]; for directed graphs with self-connections allowed, [math]\displaystyle{ E_{\max}=N^2 }[/math]. In the circumstance of a graph within which multiple edges may exist between a pair of vertices, [math]\displaystyle{ E_{\max}=\infty }[/math].
Density
The density [math]\displaystyle{ D }[/math] of a network is defined as a normalized ratio between 0 and 1 of the number of edges [math]\displaystyle{ E }[/math] to the number of possible edges in a network with [math]\displaystyle{ N }[/math] nodes. Network density is a measure of the percentage of "optional" edges that exist in the network and can be computed as [math]\displaystyle{ D =\frac{E-E_{\mathrm{min}}}{E_{\mathrm{max}}-E_{\mathrm{min}}} }[/math] where [math]\displaystyle{ E_{\mathrm{min}} }[/math] and [math]\displaystyle{ E_{\mathrm{max}} }[/math] are the minimum and maximum number of edges in a connected network with [math]\displaystyle{ N }[/math] nodes, respectively. In the case of simple graphs, [math]\displaystyle{ E_{\mathrm{max}} }[/math] is given by the binomial coefficient [math]\displaystyle{ \tbinom N2 }[/math] and [math]\displaystyle{ E_{\mathrm{min}} = N-1 }[/math], giving density [math]\displaystyle{ D =\frac{E-(N-1)}{E_{\mathrm{max}} - (N-1)} = \frac{2(E-N+1)}{N(N-3)+2} }[/math]. Another possible equation is [math]\displaystyle{ D = \frac{T-2N+2}{N(N-3)+2}, }[/math] whereas the ties [math]\displaystyle{ T }[/math] are unidirectional (Wasserman & Faust 1994).[8] This gives a better overview over the network density, because unidirectional relationships can be measured.
Planar network density
The density [math]\displaystyle{ D }[/math] of a network, where there is no intersection between edges, is defined as a ratio of the number of edges [math]\displaystyle{ E }[/math] to the number of possible edges in a network with [math]\displaystyle{ N }[/math] nodes, given by a graph with no intersecting edges [math]\displaystyle{ (E_{\max}=3N-6) }[/math], giving [math]\displaystyle{ D = \frac{E-N+1}{2N-5}. }[/math]
Average degree
The degree [math]\displaystyle{ k }[/math] of a node is the number of edges connected to it. Closely related to the density of a network is the average degree, [math]\displaystyle{ \langle k\rangle = \tfrac{2E}{N} }[/math] (or, in the case of directed graphs, [math]\displaystyle{ \langle k\rangle = \tfrac{E}{N} }[/math], the former factor of 2 arising from each edge in an undirected graph contributing to the degree of two distinct vertices). In the ER random graph model ([math]\displaystyle{ G(N,p) }[/math]) we can compute the expected value of [math]\displaystyle{ \langle k \rangle }[/math] (equal to the expected value of [math]\displaystyle{ k }[/math] of an arbitrary vertex): a random vertex has [math]\displaystyle{ N-1 }[/math] other vertices in the network available, and with probability [math]\displaystyle{ p }[/math], connects to each. Thus, [math]\displaystyle{ \mathbb{E}[\langle k \rangle]=\mathbb{E}[k]= p(N-1) }[/math].
Average shortest path length (or characteristic path length)
The average shortest path length is calculated by finding the shortest path between all pairs of nodes, and taking the average over all paths of the length thereof (the length being the number of intermediate edges contained in the path, i.e., the distance [math]\displaystyle{ d_{u,v} }[/math] between the two vertices [math]\displaystyle{ u,v }[/math] within the graph). This shows us, on average, the number of steps it takes to get from one member of the network to another. The behavior of the expected average shortest path length (that is, the ensemble average of the average shortest path length) as a function of the number of vertices [math]\displaystyle{ N }[/math] of a random network model defines whether that model exhibits the small-world effect; if it scales as [math]\displaystyle{ O(\ln N) }[/math], the model generates small-world nets. For faster-than-logarithmic growth, the model does not produce small worlds. The special case of [math]\displaystyle{ O(\ln\ln N) }[/math] is known as ultra-small world effect.
Diameter of a network
As another means of measuring network graphs, we can define the diameter of a network as the longest of all the calculated shortest paths in a network. It is the shortest distance between the two most distant nodes in the network. In other words, once the shortest path length from every node to all other nodes is calculated, the diameter is the longest of all the calculated path lengths. The diameter is representative of the linear size of a network. If node A-B-C-D are connected, going from A->D this would be the diameter of 3 (3-hops, 3-links).[citation needed]
Clustering coefficient
The clustering coefficient is a measure of an "all-my-friends-know-each-other" property. This is sometimes described as the friends of my friends are my friends. More precisely, the clustering coefficient of a node is the ratio of existing links connecting a node's neighbors to each other to the maximum possible number of such links. The clustering coefficient for the entire network is the average of the clustering coefficients of all the nodes. A high clustering coefficient for a network is another indication of a small world.
The clustering coefficient of the [math]\displaystyle{ i }[/math]'th node is
- [math]\displaystyle{ C_i = {2e_i\over k_i{(k_i - 1)}}\,, }[/math]
where [math]\displaystyle{ k_i }[/math] is the number of neighbours of the [math]\displaystyle{ i }[/math]'th node, and [math]\displaystyle{ e_i }[/math] is the number of connections between these neighbours. The maximum possible number of connections between neighbors is, then,
- [math]\displaystyle{ {\binom {k}{2}} = {{k(k-1)}\over 2}\,. }[/math]
From a probabilistic standpoint, the expected local clustering coefficient is the likelihood of a link existing between two arbitrary neighbors of the same node.
Connectedness
The way in which a network is connected plays a large part into how networks are analyzed and interpreted. Networks are classified in four different categories:
- Clique/Complete Graph: a completely connected network, where all nodes are connected to every other node. These networks are symmetric in that all nodes have in-links and out-links from all others.
- Giant Component: A single connected component which contains most of the nodes in the network.
- Weakly Connected Component: A collection of nodes in which there exists a path from any node to any other, ignoring directionality of the edges.
- Strongly Connected Component: A collection of nodes in which there exists a directed path from any node to any other.
Node centrality
Centrality indices produce rankings which seek to identify the most important nodes in a network model. Different centrality indices encode different contexts for the word "importance." The betweenness centrality, for example, considers a node highly important if it form bridges between many other nodes. The eigenvalue centrality, in contrast, considers a node highly important if many other highly important nodes link to it. Hundreds of such measures have been proposed in the literature.
Centrality indices are only accurate for identifying the most important nodes. The measures are seldom, if ever, meaningful for the remainder of network nodes.[9][10] Also, their indications are only accurate within their assumed context for importance, and tend to "get it wrong" for other contexts.[11] For example, imagine two separate communities whose only link is an edge between the most junior member of each community. Since any transfer from one community to the other must go over this link, the two junior members will have high betweenness centrality. But, since they are junior, (presumably) they have few connections to the "important" nodes in their community, meaning their eigenvalue centrality would be quite low.
Node influence
Limitations to centrality measures have led to the development of more general measures. Two examples are the accessibility, which uses the diversity of random walks to measure how accessible the rest of the network is from a given start node,[12] and the expected force, derived from the expected value of the force of infection generated by a node.[9] Both of these measures can be meaningfully computed from the structure of the network alone.
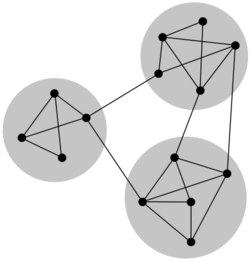
Community structure
Nodes in a network may be partitioned into groups representing communities. Depending on the context, communities may be distinct or overlapping. Typically, nodes in such communities will be strongly connected to other nodes in the same community, but weakly connected to nodes outside the community. In the absence of a ground truth describing the community structure of a specific network, several algorithms have been developed to infer possible community structures using either supervised of unsupervised clustering methods.
Network models
Network models serve as a foundation to understanding interactions within empirical complex networks. Various random graph generation models produce network structures that may be used in comparison to real-world complex networks.
Erdős–Rényi random graph model
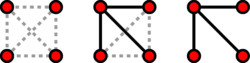
The Erdős–Rényi model, named for Paul Erdős and Alfréd Rényi, is used for generating random graphs in which edges are set between nodes with equal probabilities. It can be used in the probabilistic method to prove the existence of graphs satisfying various properties, or to provide a rigorous definition of what it means for a property to hold for almost all graphs.
To generate an Erdős–Rényi model [math]\displaystyle{ G(n, p) }[/math] two parameters must be specified: the total number of nodes n and the probability p that a random pair of nodes has an edge.
Because the model is generated without bias to particular nodes, the degree distribution is binomial: for a randomly chosen vertex [math]\displaystyle{ v }[/math],
- [math]\displaystyle{ P(\deg(v) = k) = {n-1\choose k} p^k (1-p)^{n-1-k}. }[/math]
In this model the clustering coefficient is 0 a.s. The behavior of [math]\displaystyle{ G(n, p) }[/math] can be broken into three regions.
Subcritical [math]\displaystyle{ n p \lt 1 }[/math]: All components are simple and very small, the largest component has size [math]\displaystyle{ |C_1| = O(\log n) }[/math];
Critical [math]\displaystyle{ n p = 1 }[/math]: [math]\displaystyle{ |C_1| = O(n^\frac{2}{3}) }[/math];
Supercritical [math]\displaystyle{ n p \gt 1 }[/math]:[math]\displaystyle{ |C_1| \approx yn }[/math] where [math]\displaystyle{ y = y(n p) }[/math] is the positive solution to the equation [math]\displaystyle{ e^{-p n y }=1-y }[/math].
The largest connected component has high complexity. All other components are simple and small [math]\displaystyle{ |C_2| = O(\log n) }[/math].
Configuration model
The configuration model takes a degree sequence[13][14] or degree distribution[15] (which subsequently is used to generate a degree sequence) as the input, and produces randomly connected graphs in all respects other than the degree sequence. This means that for a given choice of the degree sequence, the graph is chosen uniformly at random from the set of all graphs that comply with this degree sequence. The degree [math]\displaystyle{ k }[/math] of a randomly chosen vertex is an independent and identically distributed random variable with integer values. When [math]\displaystyle{ \mathbb E [k^2] - 2 \mathbb E [k]\gt 0 }[/math], the configuration graph contains the giant connected component, which has infinite size.[14] The rest of the components have finite sizes, which can be quantified with the notion of the size distribution. The probability [math]\displaystyle{ w(n) }[/math] that a randomly sampled node is connected to a component of size [math]\displaystyle{ n }[/math] is given by convolution powers of the degree distribution:[16][math]\displaystyle{ w(n)=\begin{cases} \frac{\mathbb E [k]}{n-1} u_1^{*n}(n-2),& n\gt 1, \\ u(0) & n=1, \end{cases} }[/math]where [math]\displaystyle{ u(k) }[/math] denotes the degree distribution and [math]\displaystyle{ u_1(k)=\frac{(k+1)u(k+1)}{\mathbb E[k]} }[/math]. The giant component can be destroyed by randomly removing the critical fraction [math]\displaystyle{ p_c }[/math] of all edges. This process is called percolation on random networks. When the second moment of the degree distribution is finite, [math]\displaystyle{ \mathbb E [k^2]\lt \infty }[/math], this critical edge fraction is given by[17] [math]\displaystyle{ p_c=1-\frac{\mathbb E[k]}{ \mathbb E [k^2] - \mathbb E[k]} }[/math], and the average vertex-vertex distance [math]\displaystyle{ l }[/math] in the giant component scales logarithmically with the total size of the network, [math]\displaystyle{ l = O(\log N) }[/math].[15]
In the directed configuration model, the degree of a node is given by two numbers, in-degree [math]\displaystyle{ k_\text{in} }[/math] and out-degree [math]\displaystyle{ k_\text{out} }[/math], and consequently, the degree distribution is two-variate. The expected number of in-edges and out-edges coincides, so that [math]\displaystyle{ \mathbb E [k_\text{in}]=\mathbb E [k_\text{out}] }[/math]. The directed configuration model contains the giant component iff[18][math]\displaystyle{ 2\mathbb{E}[k_\text{in}] \mathbb{E}[k_\text{in}k_\text{out}] - \mathbb{E}[k_\text{in}] \mathbb{E}[k_\text{out}^2] - \mathbb{E}[k_\text{in}] \mathbb{E}[k_\text{in}^2] +\mathbb{E}[k_\text{in}^2] \mathbb{E}[k_\text{out}^2] - \mathbb{E}[k_\text{in} k_\text{out}]^2 \gt 0. }[/math]Note that [math]\displaystyle{ \mathbb E [k_\text{in}] }[/math] and [math]\displaystyle{ \mathbb E [k_\text{out}] }[/math] are equal and therefore interchangeable in the latter inequality. The probability that a randomly chosen vertex belongs to a component of size [math]\displaystyle{ n }[/math] is given by:[19][math]\displaystyle{ h_\text{in}(n)=\frac{\mathbb E[k_{in}]}{n-1} \tilde u_\text{in}^{*n}(n-2), \;n\gt 1, \; \tilde u_\text{in}=\frac{k_\text{in}+1}{\mathbb E[k_\text{in}]}\sum\limits_{k_\text{out}\geq 0}u(k_\text{in}+1,k_\text{out}), }[/math]for in-components, and
- [math]\displaystyle{ h_\text{out}(n)=\frac{\mathbb E[k_\text{out}]}{n-1} \tilde u_\text{out}^{*n}(n-2), \;n\gt 1, \;\tilde u_\text{out}=\frac{k_\text{out}+1}{\mathbb E[k_\text{out}]}\sum\limits_{k_\text{in}\geq0}u(k_\text{in},k_\text{out}+1), }[/math]
for out-components.
Watts–Strogatz small world model
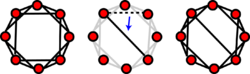
The Watts and Strogatz model is a random graph generation model that produces graphs with small-world properties.
An initial lattice structure is used to generate a Watts–Strogatz model. Each node in the network is initially linked to its [math]\displaystyle{ \langle k\rangle }[/math] closest neighbors. Another parameter is specified as the rewiring probability. Each edge has a probability [math]\displaystyle{ p }[/math] that it will be rewired to the graph as a random edge. The expected number of rewired links in the model is [math]\displaystyle{ pE = pN\langle k\rangle/2 }[/math].
As the Watts–Strogatz model begins as non-random lattice structure, it has a very high clustering coefficient along with high average path length. Each rewire is likely to create a shortcut between highly connected clusters. As the rewiring probability increases, the clustering coefficient decreases slower than the average path length. In effect, this allows the average path length of the network to decrease significantly with only slightly decreases in clustering coefficient. Higher values of p force more rewired edges, which in effect makes the Watts–Strogatz model a random network.
Barabási–Albert (BA) preferential attachment model
The Barabási–Albert model is a random network model used to demonstrate a preferential attachment or a "rich-get-richer" effect. In this model, an edge is most likely to attach to nodes with higher degrees. The network begins with an initial network of m0 nodes. m0 ≥ 2 and the degree of each node in the initial network should be at least 1, otherwise it will always remain disconnected from the rest of the network.
In the BA model, new nodes are added to the network one at a time. Each new node is connected to [math]\displaystyle{ m }[/math] existing nodes with a probability that is proportional to the number of links that the existing nodes already have. Formally, the probability pi that the new node is connected to node i is[20]
- [math]\displaystyle{ p_i = \frac{k_i}{\sum_j k_j}, }[/math]
where ki is the degree of node i. Heavily linked nodes ("hubs") tend to quickly accumulate even more links, while nodes with only a few links are unlikely to be chosen as the destination for a new link. The new nodes have a "preference" to attach themselves to the already heavily linked nodes.

The degree distribution resulting from the BA model is scale free, in particular, for large degree it is a power law of the form:
- [math]\displaystyle{ P(k)\sim k^{-3} \, }[/math]
Hubs exhibit high betweenness centrality which allows short paths to exist between nodes. As a result, the BA model tends to have very short average path lengths. The clustering coefficient of this model also tends to 0.
The Barabási–Albert model[21] was developed for undirected networks but applied to a wide range of different applications. The directed version of this model is the Price model[22][23] which was applied to just citation networks. Mathematically, the same equations can be used to describe these models[24] and they both show the same features.
Non-linear preferential attachment
In non-linear preferential attachment (NLPA), existing nodes in the network gain new edges proportionally to the node degree raised to a constant positive power, [math]\displaystyle{ \alpha }[/math].[25] Formally, this means that the probability that node [math]\displaystyle{ i }[/math] gains a new edge is given by
- [math]\displaystyle{ p_i = \frac{k_i^{\alpha}}{\sum_j k_j^{\alpha}}. }[/math]
If [math]\displaystyle{ \alpha=1 }[/math], NLPA reduces to the BA model and is referred to as "linear". If [math]\displaystyle{ 0\lt \alpha\lt 1 }[/math], NLPA is referred to as "sub-linear" and the degree distribution of the network tends to a stretched exponential distribution. If [math]\displaystyle{ \alpha\gt 1 }[/math], NLPA is referred to as "super-linear" and a small number of nodes connect to almost all other nodes in the network. For both [math]\displaystyle{ \alpha\lt 1 }[/math] and [math]\displaystyle{ \alpha\gt 1 }[/math], the scale-free property of the network is broken in the limit of infinite system size. However, if [math]\displaystyle{ \alpha }[/math] is only slightly larger than [math]\displaystyle{ 1 }[/math], NLPA may result in degree distributions which appear to be transiently scale free.[26]
Mediation-driven attachment (MDA) model
In the mediation-driven attachment (MDA) model in which a new node coming with [math]\displaystyle{ m }[/math] edges picks an existing connected node at random and then connects itself not with that one but with [math]\displaystyle{ m }[/math] of its neighbors chosen also at random. The probability [math]\displaystyle{ \Pi(i) }[/math] that the node [math]\displaystyle{ i }[/math] of the existing node picked is
- [math]\displaystyle{ \Pi(i) = \frac{k_i} N \frac{ \sum_{j=1}^{k_i} \frac 1 {k_j} }{k_i}. }[/math]
The factor [math]\displaystyle{ \frac{\sum_{j=1}^{k_i}{\frac{1}{k_j}}}{k_i} }[/math] is the inverse of the harmonic mean (IHM) of degrees of the [math]\displaystyle{ k_i }[/math] neighbors of a node [math]\displaystyle{ i }[/math]. Extensive numerical investigation suggest that for an approximately [math]\displaystyle{ m\gt 14 }[/math] the mean IHM value in the large [math]\displaystyle{ N }[/math] limit becomes a constant which means [math]\displaystyle{ \Pi(i) \propto k_i }[/math]. It implies that the higher the links (degree) a node has, the higher its chance of gaining more links since they can be reached in a larger number of ways through mediators which essentially embodies the intuitive idea of rich get richer mechanism (or the preferential attachment rule of the Barabasi–Albert model). Therefore, the MDA network can be seen to follow the PA rule but in disguise.[27]
However, for [math]\displaystyle{ m=1 }[/math] it describes the winner takes it all mechanism as we find that almost [math]\displaystyle{ 99\% }[/math] of the total nodes have degree one and one is super-rich in degree. As [math]\displaystyle{ m }[/math] value increases the disparity between the super rich and poor decreases and as [math]\displaystyle{ m\gt 14 }[/math] we find a transition from rich get super richer to rich get richer mechanism.
Fitness model
Another model where the key ingredient is the nature of the vertex has been introduced by Caldarelli et al.[28] Here a link is created between two vertices [math]\displaystyle{ i,j }[/math] with a probability given by a linking function [math]\displaystyle{ f(\eta_i,\eta_j) }[/math] of the fitnesses of the vertices involved. The degree of a vertex i is given by [29]
- [math]\displaystyle{ k(\eta_i)=N\int_0^\infty f(\eta_i,\eta_j) \rho(\eta_j) \, d\eta_j }[/math]
If [math]\displaystyle{ k(\eta_i) }[/math] is an invertible and increasing function of [math]\displaystyle{ \eta_i }[/math], then the probability distribution [math]\displaystyle{ P(k) }[/math] is given by
- [math]\displaystyle{ P(k)=\rho(\eta(k)) \cdot \eta'(k) }[/math]
As a result, if the fitnesses [math]\displaystyle{ \eta }[/math] are distributed as a power law, then also the node degree does.
Less intuitively with a fast decaying probability distribution as [math]\displaystyle{ \rho(\eta)=e^{-\eta} }[/math] together with a linking function of the kind
- [math]\displaystyle{ f(\eta_i,\eta_j)=\Theta(\eta_i+\eta_j-Z) }[/math]
with [math]\displaystyle{ Z }[/math] a constant and [math]\displaystyle{ \Theta }[/math] the Heavyside function, we also obtain scale-free networks.
Such model has been successfully applied to describe trade between nations by using GDP as fitness for the various nodes [math]\displaystyle{ i,j }[/math] and a linking function of the kind [30][31]
- [math]\displaystyle{ \frac{\delta \eta_i\eta_j}{1+ \delta \eta_i\eta_j}. }[/math]
Exponential random graph models
Exponential Random Graph Models (ERGMs) are a family of statistical models for analyzing data from social and other networks.[32] The Exponential family is a broad family of models for covering many types of data, not just networks. An ERGM is a model from this family which describes networks.
We adopt the notation to represent a random graph [math]\displaystyle{ Y \in \mathcal{Y} }[/math] via a set of [math]\displaystyle{ n }[/math] nodes and a collection of tie variables [math]\displaystyle{ \{ Y_{ij}: i=1,\dots,n; j=1,\dots,n\} }[/math], indexed by pairs of nodes [math]\displaystyle{ ij }[/math], where [math]\displaystyle{ Y_{ij}=1 }[/math] if the nodes [math]\displaystyle{ (i,j) }[/math] are connected by an edge and [math]\displaystyle{ Y_{ij}=0 }[/math] otherwise.
The basic assumption of ERGMs is that the structure in an observed graph [math]\displaystyle{ y }[/math] can be explained by a given vector of sufficient statistics [math]\displaystyle{ s(y) }[/math] which are a function of the observed network and, in some cases, nodal attributes. The probability of a graph [math]\displaystyle{ y\in\mathcal{Y} }[/math] in an ERGM is defined by:
[math]\displaystyle{ P(Y = y | \theta) = \frac{\exp(\theta^{T} s(y))}{c(\theta)} }[/math]
where [math]\displaystyle{ \theta }[/math] is a vector of model parameters associated with [math]\displaystyle{ s(y) }[/math] and [math]\displaystyle{ c(\theta) = \sum_{y'\in\mathcal{Y}}\exp(\theta^{T} s(y')) }[/math] is a normalising constant.
Network analysis
Social network analysis
Social network analysis examines the structure of relationships between social entities.[33] These entities are often persons, but may also be groups, organizations, nation states, web sites, scholarly publications.
Since the 1970s, the empirical study of networks has played a central role in social science, and many of the mathematical and statistical tools used for studying networks have been first developed in sociology.[24] Amongst many other applications, social network analysis has been used to understand the diffusion of innovations,[34] news and rumors. Similarly, it has been used to examine the spread of both diseases and health-related behaviors. It has also been applied to the study of markets, where it has been used to examine the role of trust in exchange relationships and of social mechanisms in setting prices. Similarly, it has been used to study recruitment into political movements and social organizations. It has also been used to conceptualize scientific disagreements as well as academic prestige. In the second language acquisition literature, it has an established history in study abroad research, revealing how peer learner interaction networks influence their language progress.[35] More recently, network analysis (and its close cousin traffic analysis) has gained a significant use in military intelligence, for uncovering insurgent networks of both hierarchical and leaderless nature.[36][37] In criminology, it is being used to identify influential actors in criminal gangs, offender movements, co-offending, predict criminal activities and make policies.[38]
Dynamic network analysis
Dynamic network analysis examines the shifting structure of relationships among different classes of entities in complex socio-technical systems effects, and reflects social stability and changes such as the emergence of new groups, topics, and leaders.[39][40][41] Dynamic Network Analysis focuses on meta-networks composed of multiple types of nodes (entities) and multiple types of links. These entities can be highly varied. Examples include people, organizations, topics, resources, tasks, events, locations, and beliefs.
Dynamic network techniques are particularly useful for assessing trends and changes in networks over time, identification of emergent leaders, and examining the co-evolution of people and ideas.
Biological network analysis
With the recent explosion of publicly available high throughput biological data, the analysis of molecular networks has gained significant interest. The type of analysis in this content are closely related to social network analysis, but often focusing on local patterns in the network. For example, network motifs are small subgraphs that are over-represented in the network. Activity motifs are similar over-represented patterns in the attributes of nodes and edges in the network that are over represented given the network structure. The analysis of biological networks has led to the development of network medicine, which looks at the effect of diseases in the interactome.[42]
Link analysis
Link analysis is a subset of network analysis, exploring associations between objects. An example may be examining the addresses of suspects and victims, the telephone numbers they have dialed and financial transactions that they have partaken in during a given timeframe, and the familial relationships between these subjects as a part of police investigation. Link analysis here provides the crucial relationships and associations between very many objects of different types that are not apparent from isolated pieces of information. Computer-assisted or fully automatic computer-based link analysis is increasingly employed by banks and insurance agencies in fraud detection, by telecommunication operators in telecommunication network analysis, by medical sector in epidemiology and pharmacology, in law enforcement investigations, by search engines for relevance rating (and conversely by the spammers for spamdexing and by business owners for search engine optimization), and everywhere else where relationships between many objects have to be analyzed.
Pandemic analysis
The SIR model is one of the most well known algorithms on predicting the spread of global pandemics within an infectious population.
Susceptible to infected
- [math]\displaystyle{ S = \beta\left(\frac 1 N \right) }[/math]
The formula above describes the "force" of infection for each susceptible unit in an infectious population, where β is equivalent to the transmission rate of said disease.
To track the change of those susceptible in an infectious population:
- [math]\displaystyle{ \Delta S = \beta \times S {1\over N} \, \Delta t }[/math]
Infected to recovered
- [math]\displaystyle{ \Delta I = \mu I \, \Delta t }[/math]
Over time, the number of those infected fluctuates by: the specified rate of recovery, represented by [math]\displaystyle{ \mu }[/math] but deducted to one over the average infectious period [math]\displaystyle{ {1\over \tau} }[/math], the numbered of infectious individuals, [math]\displaystyle{ I }[/math], and the change in time, [math]\displaystyle{ \Delta t }[/math].
Infectious period
Whether a population will be overcome by a pandemic, with regards to the SIR model, is dependent on the value of [math]\displaystyle{ R_0 }[/math] or the "average people infected by an infected individual."
- [math]\displaystyle{ R_0 = \beta\tau = {\beta\over\mu} }[/math]
Web link analysis
Several Web search ranking algorithms use link-based centrality metrics, including (in order of appearance) Marchiori's Hyper Search, Google's PageRank, Kleinberg's HITS algorithm, the CheiRank and TrustRank algorithms. Link analysis is also conducted in information science and communication science in order to understand and extract information from the structure of collections of web pages. For example, the analysis might be of the interlinking between politicians' web sites or blogs.
PageRank
PageRank works by randomly picking "nodes" or websites and then with a certain probability, "randomly jumping" to other nodes. By randomly jumping to these other nodes, it helps PageRank completely traverse the network as some webpages exist on the periphery and would not as readily be assessed.
Each node, [math]\displaystyle{ x_i }[/math], has a PageRank as defined by the sum of pages [math]\displaystyle{ j }[/math] that link to [math]\displaystyle{ i }[/math] times one over the outlinks or "out-degree" of [math]\displaystyle{ j }[/math] times the "importance" or PageRank of [math]\displaystyle{ j }[/math].
- [math]\displaystyle{ x_i = \sum_{j\rightarrow i}{1\over N_j}x_j^{(k)} }[/math]
Random jumping
As explained above, PageRank enlists random jumps in attempts to assign PageRank to every website on the internet. These random jumps find websites that might not be found during the normal search methodologies such as breadth-first search and depth-first search.
In an improvement over the aforementioned formula for determining PageRank includes adding these random jump components. Without the random jumps, some pages would receive a PageRank of 0 which would not be good.
The first is [math]\displaystyle{ \alpha }[/math], or the probability that a random jump will occur. Contrasting is the "damping factor", or [math]\displaystyle{ 1 - \alpha }[/math].
- [math]\displaystyle{ R{(p)} = {\alpha\over N} + (1 - \alpha) \sum_{j\rightarrow i} {1\over N_j} x_j^{(k)} }[/math]
Another way of looking at it:
- [math]\displaystyle{ R(A) = \sum {R_B\over B_\text{(outlinks)}} + \cdots + {R_n \over n_\text{(outlinks)}} }[/math]
Centrality measures
Information about the relative importance of nodes and edges in a graph can be obtained through centrality measures, widely used in disciplines like sociology. Centrality measures are essential when a network analysis has to answer questions such as: "Which nodes in the network should be targeted to ensure that a message or information spreads to all or most nodes in the network?" or conversely, "Which nodes should be targeted to curtail the spread of a disease?". Formally established measures of centrality are degree centrality, closeness centrality, betweenness centrality, eigenvector centrality, and katz centrality. The objective of network analysis generally determines the type of centrality measure(s) to be used.[33]
- Degree centrality of a node in a network is the number of links (vertices) incident on the node.
- Closeness centrality determines how "close" a node is to other nodes in a network by measuring the sum of the shortest distances (geodesic paths) between that node and all other nodes in the network.
- Betweenness centrality determines the relative importance of a node by measuring the amount of traffic flowing through that node to other nodes in the network. This is done by measuring the fraction of paths connecting all pairs of nodes and containing the node of interest. Group Betweenness centrality measures the amount of traffic flowing through a group of nodes.
- Eigenvector centrality is a more sophisticated version of degree centrality where the centrality of a node not only depends on the number of links incident on the node but also the quality of those links. This quality factor is determined by the eigenvectors of the adjacency matrix of the network.
- Katz centrality of a node is measured by summing the geodesic paths between that node and all (reachable) nodes in the network. These paths are weighted, paths connecting the node with its immediate neighbors carry higher weights than those which connect with nodes farther away from the immediate neighbors.
Spread of content in networks
Content in a complex network can spread via two major methods: conserved spread and non-conserved spread.[43] In conserved spread, the total amount of content that enters a complex network remains constant as it passes through. The model of conserved spread can best be represented by a pitcher containing a fixed amount of water being poured into a series of funnels connected by tubes. Here, the pitcher represents the original source and the water is the content being spread. The funnels and connecting tubing represent the nodes and the connections between nodes, respectively. As the water passes from one funnel into another, the water disappears instantly from the funnel that was previously exposed to the water. In non-conserved spread, the amount of content changes as it enters and passes through a complex network. The model of non-conserved spread can best be represented by a continuously running faucet running through a series of funnels connected by tubes. Here, the amount of water from the original source is infinite. Also, any funnels that have been exposed to the water continue to experience the water even as it passes into successive funnels. The non-conserved model is the most suitable for explaining the transmission of most infectious diseases.
The SIR model
In 1927, W. O. Kermack and A. G. McKendrick created a model in which they considered a fixed population with only three compartments, susceptible: [math]\displaystyle{ S(t) }[/math], infected, [math]\displaystyle{ I(t) }[/math], and recovered, [math]\displaystyle{ R(t) }[/math]. The compartments used for this model consist of three classes:
- [math]\displaystyle{ S(t) }[/math] is used to represent the number of individuals not yet infected with the disease at time t, or those susceptible to the disease
- [math]\displaystyle{ I(t) }[/math] denotes the number of individuals who have been infected with the disease and are capable of spreading the disease to those in the susceptible category
- [math]\displaystyle{ R(t) }[/math] is the compartment used for those individuals who have been infected and then recovered from the disease. Those in this category are not able to be infected again or to transmit the infection to others.
The flow of this model may be considered as follows:
- [math]\displaystyle{ \mathcal{S} \rightarrow \mathcal{I} \rightarrow \mathcal{R} }[/math]
Using a fixed population, [math]\displaystyle{ N = S(t) + I(t) + R(t) }[/math], Kermack and McKendrick derived the following equations:
- [math]\displaystyle{ \begin{align} \frac{dS}{dt} & = - \beta S I \\[8pt] \frac{dI}{dt} & = \beta S I - \gamma I \\[8pt] \frac{dR}{dt} & = \gamma I \end{align} }[/math]
Several assumptions were made in the formulation of these equations: First, an individual in the population must be considered as having an equal probability as every other individual of contracting the disease with a rate of [math]\displaystyle{ \beta }[/math], which is considered the contact or infection rate of the disease. Therefore, an infected individual makes contact and is able to transmit the disease with [math]\displaystyle{ \beta N }[/math] others per unit time and the fraction of contacts by an infected with a susceptible is [math]\displaystyle{ S/N }[/math]. The number of new infections in unit time per infective then is [math]\displaystyle{ \beta N (S/N) }[/math], giving the rate of new infections (or those leaving the susceptible category) as [math]\displaystyle{ \beta N (S/N)I = \beta SI }[/math] (Brauer & Castillo-Chavez, 2001). For the second and third equations, consider the population leaving the susceptible class as equal to the number entering the infected class. However, infectives are leaving this class per unit time to enter the recovered/removed class at a rate [math]\displaystyle{ \gamma }[/math] per unit time (where [math]\displaystyle{ \gamma }[/math] represents the mean recovery rate, or [math]\displaystyle{ 1/\gamma }[/math] the mean infective period). These processes which occur simultaneously are referred to as the Law of Mass Action, a widely accepted idea that the rate of contact between two groups in a population is proportional to the size of each of the groups concerned (Daley & Gani, 2005). Finally, it is assumed that the rate of infection and recovery is much faster than the time scale of births and deaths and therefore, these factors are ignored in this model.
More can be read on this model on the Epidemic model page.
The master equation approach
A master equation can express the behaviour of an undirected growing network where, at each time step, a new node is added to the network, linked to an old node (randomly chosen and without preference). The initial network is formed by two nodes and two links between them at time [math]\displaystyle{ t = 2 }[/math], this configuration is necessary only to simplify further calculations, so at time [math]\displaystyle{ t = n }[/math] the network have [math]\displaystyle{ n }[/math] nodes and [math]\displaystyle{ n }[/math] links.
The master equation for this network is:
- [math]\displaystyle{ p(k,s,t+1) = \frac 1 t p(k-1,s,t) + \left(1 - \frac 1 t \right)p(k,s,t), }[/math]
where [math]\displaystyle{ p(k,s,t) }[/math] is the probability to have the node [math]\displaystyle{ s }[/math] with degree [math]\displaystyle{ k }[/math] at time [math]\displaystyle{ t+1 }[/math], and [math]\displaystyle{ s }[/math] is the time step when this node was added to the network. Note that there are only two ways for an old node [math]\displaystyle{ s }[/math] to have [math]\displaystyle{ k }[/math] links at time [math]\displaystyle{ t+1 }[/math]:
- The node [math]\displaystyle{ s }[/math] have degree [math]\displaystyle{ k-1 }[/math] at time [math]\displaystyle{ t }[/math] and will be linked by the new node with probability [math]\displaystyle{ 1/t }[/math]
- Already has degree [math]\displaystyle{ k }[/math] at time [math]\displaystyle{ t }[/math] and will not be linked by the new node.
After simplifying this model, the degree distribution is [math]\displaystyle{ P(k) = 2^{-k}. }[/math][44]
Based on this growing network, an epidemic model is developed following a simple rule: Each time the new node is added and after choosing the old node to link, a decision is made: whether or not this new node will be infected. The master equation for this epidemic model is:
- [math]\displaystyle{ p_r(k,s,t) = r_t \frac 1 t p_r(k-1,s,t) + \left(1 - \frac 1 t \right) p_r(k,s,t), }[/math]
where [math]\displaystyle{ r_t }[/math] represents the decision to infect ([math]\displaystyle{ r_t = 1 }[/math]) or not ([math]\displaystyle{ r_t = 0 }[/math]). Solving this master equation, the following solution is obtained: [math]\displaystyle{ \tilde{P}_r(k) = \left(\frac r 2 \right)^k. }[/math][45]
Multilayer networks
Multilayer networks are networks with multiple kinds of relations.[46] Attempts to model real-world systems as multidimensional networks have been used in various fields such as social network analysis,[47] economics, history, urban and international transport, ecology, psychology, medicine, biology, commerce, climatology, physics, computational neuroscience, operations management, and finance.
Network optimization
Network problems that involve finding an optimal way of doing something are studied under the name of combinatorial optimization. Examples include network flow, shortest path problem, transport problem, transshipment problem, location problem, matching problem, assignment problem, packing problem, routing problem, critical path analysis and PERT (Program Evaluation & Review Technique).
Interdependent networks
Interdependent networks are networks where the functioning of nodes in one network depends on the functioning of nodes in another network. In nature, networks rarely appear in isolation, rather, usually networks are typically elements in larger systems, and interact with elements in that complex system. Such complex dependencies can have non-trivial effects on one another. A well studied example is the interdependency of infrastructure networks,[48] the power stations which form the nodes of the power grid require fuel delivered via a network of roads or pipes and are also controlled via the nodes of communications network. Though the transportation network does not depend on the power network to function, the communications network does. In such infrastructure networks, the disfunction of a critical number of nodes in either the power network or the communication network can lead to cascading failures across the system with potentially catastrophic result to the whole system functioning.[49] If the two networks were treated in isolation, this important feedback effect would not be seen and predictions of network robustness would be greatly overestimated.
See also
- Cascading failure
- Climate as complex networks
- Collaborative innovation network
- Communicative ecology
- Complex network
- Core-periphery structures in networks
- Dual-phase evolution
- Erdős–Rényi model
- Glossary of graph theory
- Gradient network
- Higher category theory
- Immune network theory
- Irregular warfare
- Network analyzer
- Network dynamics
- Network formation
- Network theory in risk assessment
- Network topology
- Networks in labor economics
- Non-linear preferential attachment
- Percolation
- Percolation theory
- Policy network analysis
- Polytely
- Quantum complex network
- Random networks
- Rumor spread in social network
- Scale-free networks
- Sequential dynamical system
- Service network
- Small-world networks
- Structural cut-off
- Systems theory
References
- ↑ Committee on Network Science for Future Army Applications (2006). Network Science. National Research Council. doi:10.17226/11516. ISBN 978-0309653886. http://www.nap.edu/catalog/11516.html.
- ↑ Dénes Kőnig (1990). Theory of finite and infinite graphs (PDF). Birkhäuser Boston. pp. 45–421. doi:10.1007/978-1-4684-8971-2. ISBN 978-1-4684-8971-2. https://link.springer.com/content/pdf/10.1007/978-1-4684-8971-2_2.pdf.
- ↑ "Connected: The Power of Six Degrees (TV Movie 2008) - IMDb". https://www.imdb.com/title/tt1310375/fullcredits?ref_=tt_ov_st_sm#cast.
- ↑ http://citeseerx.ist.psu.edu/viewdoc/summary?doi=10.1.1.301.5111
- ↑ "Network Science at West Point: The early years". https://www.youtube.com/watch?v=zfNpKMab5eU&ab_channel=nunyourbusiness.
- ↑ https://www.westpoint.edu/academics/academic-departments/mathematical-sciences/network-science
- ↑ Kollios, George (2011-12-06). "Clustering Large Probabilistic Graphs". IEEE Transactions on Knowledge and Data Engineering 25 (2): 325–336. doi:10.1109/TKDE.2011.243. PMID 13188797.
- ↑ "APA PsycNet". http://psycnet.apa.org/journals/prs/9/4/172/.
- ↑ 9.0 9.1 Lawyer, Glenn (March 2015). "Understanding the spreading power of all nodes in a network". Scientific Reports 5 (O8665): 8665. doi:10.1038/srep08665. PMID 25727453. Bibcode: 2015NatSR...5E8665L.
- ↑ Sikic, Mile; Lancic, Alen; Antulov-Fantulin, Nino; Stefancic, Hrvoje (October 2013). "Epidemic centrality – is there an underestimated epidemic impact of network peripheral nodes?". European Physical Journal B 86 (10): 440. doi:10.1140/epjb/e2013-31025-5. Bibcode: 2013EPJB...86..440S.
- ↑ Borgatti, Stephen P. (2005). "Centrality and Network Flow". Social Networks 27: 55–71. doi:10.1016/j.socnet.2004.11.008.
- ↑ Travençolo, B. A. N.; da F. Costa, L. (2008). "Accessibility in complex networks". Physics Letters A 373 (1): 89–95. doi:10.1016/j.physleta.2008.10.069. Bibcode: 2008PhLA..373...89T.
- ↑ Bender, Edward A; Canfield, E.Rodney (May 1978). "The asymptotic number of labeled graphs with given degree sequences". Journal of Combinatorial Theory, Series A 24 (3): 296–307. doi:10.1016/0097-3165(78)90059-6. ISSN 0097-3165.
- ↑ 14.0 14.1 Molloy, Michael; Reed, Bruce (March 1995). "A critical point for random graphs with a given degree sequence" (in en). Random Structures & Algorithms 6 (2–3): 161–180. doi:10.1002/rsa.3240060204. ISSN 1042-9832.
- ↑ 15.0 15.1 Newman, M. E. J.; Strogatz, S. H.; Watts, D. J. (2001-07-24). "Random graphs with arbitrary degree distributions and their applications". Physical Review E 64 (2): 026118. doi:10.1103/PhysRevE.64.026118. PMID 11497662. Bibcode: 2001PhRvE..64b6118N.
- ↑ Kryven, Ivan (2017-05-02). "General expression for the component size distribution in infinite configuration networks". Physical Review E 95 (5): 052303. doi:10.1103/PhysRevE.95.052303. PMID 28618550. Bibcode: 2017PhRvE..95e2303K.
- ↑ Kryven, Ivan (2018-01-01). "Analytic results on the polymerisation random graph model" (in en). Journal of Mathematical Chemistry 56 (1): 140–157. doi:10.1007/s10910-017-0785-1. ISSN 0259-9791.
- ↑ Kryven, Ivan (2016-07-27). "Emergence of the giant weak component in directed random graphs with arbitrary degree distributions". Physical Review E 94 (1): 012315. doi:10.1103/PhysRevE.94.012315. PMID 27575156. Bibcode: 2016PhRvE..94a2315K.
- ↑ Kryven, Ivan (2017-11-02). "Finite connected components in infinite directed and multiplex networks with arbitrary degree distributions". Physical Review E 96 (5): 052304. doi:10.1103/PhysRevE.96.052304. PMID 29347790. Bibcode: 2017PhRvE..96e2304K.
- ↑ R. Albert; A.-L. Barabási (2002). "Statistical mechanics of complex networks". Reviews of Modern Physics 74 (1): 47–97. doi:10.1103/RevModPhys.74.47. Bibcode: 2002RvMP...74...47A. http://www.nd.edu/~networks/Publication%20Categories/03%20Journal%20Articles/Physics/StatisticalMechanics_Rev%20of%20Modern%20Physics%2074,%2047%20(2002).pdf.
- ↑ 21.0 21.1 Albert-László Barabási & Réka Albert (October 1999). "Emergence of scaling in random networks". Science 286 (5439): 509–512. doi:10.1126/science.286.5439.509. PMID 10521342. Bibcode: 1999Sci...286..509B. http://www.nd.edu/~networks/Publication%20Categories/03%20Journal%20Articles/Physics/EmergenceRandom_Science%20286,%20509-512%20(1999).pdf.
- ↑ Price, Derek J. de Solla (1965-07-30). "Networks of Scientific Papers: The pattern of bibliographic references indicates the nature of the scientific research front." (in en). Science 149 (3683): 510–515. doi:10.1126/science.149.3683.510. ISSN 0036-8075. PMID 14325149. Bibcode: 1965Sci...149..510D. https://www.science.org/doi/10.1126/science.149.3683.510.
- ↑ Price, Derek De Solla (1976). "A general theory of bibliometric and other cumulative advantage processes" (in en). Journal of the American Society for Information Science 27 (5): 292–306. doi:10.1002/asi.4630270505. https://onlinelibrary.wiley.com/doi/10.1002/asi.4630270505.
- ↑ 24.0 24.1 Newman, M.E.J. Networks: An Introduction. Oxford University Press. 2010, ISBN:978-0199206650
- ↑ Krapivsky, P. L.; Redner, S.; Leyvraz, F. (20 November 2000). "Connectivity of Growing Random Networks". Physical Review Letters 85 (21): 4629–4632. doi:10.1103/PhysRevLett.85.4629. PMID 11082613. Bibcode: 2000PhRvL..85.4629K.
- ↑ Krapivsky, Paul; Krioukov, Dmitri (21 August 2008). "Scale-free networks as preasymptotic regimes of superlinear preferential attachment". Physical Review E 78 (2): 026114. doi:10.1103/PhysRevE.78.026114. PMID 18850904. Bibcode: 2008PhRvE..78b6114K.
- ↑ Hassan, M. K.; Islam, Liana; Arefinul Haque, Syed (March 2017). "Degree distribution, rank-size distribution, and leadership persistence in mediation-driven attachment networks". Physica A 469: 23–30. doi:10.1016/j.physa.2016.11.001. Bibcode: 2017PhyA..469...23H.
- ↑ Caldarelli G., A. Capocci, P. De Los Rios, M.A. Muñoz, Physical Review Letters 89, 258702 (2002)
- ↑ Servedio V.D.P., G. Caldarelli, P. Buttà, Physical Review E 70, 056126 (2004)
- ↑ Garlaschelli D., M I Loffredo Physical Review Letters 93, 188701 (2004)
- ↑ Cimini G., T. Squartini, D. Garlaschelli and A. Gabrielli, Scientific Reports 5, 15758 (2015)
- ↑ Lusher, Dean; Koskinen, Johan; Robins, Garry (2012) (in English). Exponential Random Graph Models for Social Networks: Theory, Methods, and Applications (Structural Analysis in the Social Sciences). doi:10.1017/CBO9780511894701. ISBN 9780521141383. OCLC 1120539699.
- ↑ 33.0 33.1 Wasserman, Stanley and Katherine Faust. 1994. Social Network Analysis: Methods and Applications. Cambridge: Cambridge University Press.
- ↑ Paradowski, Michał B.; Jonak, Łukasz (2012). "Diffusion of linguistic innovation as social coordination". Psychology of Language and Communication 16 (2): 53–64. doi:10.2478/v10057-012-0010-z.
- ↑ Paradowski, Michał B.; Jarynowski, Andrzej; Jelińska, Magdalena; Czopek, Karolina (2021). "Out-of-class peer interactions matter for second language acquisition during short-term overseas sojourns: The contributions of Social Network Analysis [Selected poster presentations from the American Association of Applied Linguistics conference, Denver, USA, March 2020]". Language Teaching 54 (1): 139–143. doi:10.1017/S0261444820000580.
- ↑ "Toward a Complex Adaptive Intelligence Community The Wiki and the Blog". D. Calvin Andrus. cia.gov. https://www.cia.gov/library/center-for-the-study-of-intelligence/csi-publications/csi-studies/studies/vol49no3/html_files/Wik_and_%20Blog_7.htm.
- ↑ "Network analysis of terrorist networks". http://firstmonday.org/htbin/cgiwrap/bin/ojs/index.php/fm/article/view/941/863.
- ↑ PhD, Martin Bouchard; PhD, Aili Malm (2016-11-02). "Social Network Analysis and Its Contribution to Research on Crime and Criminal Justice" (in en). Oxford Handbooks Online: Criminology and Criminal Justice. doi:10.1093/oxfordhb/9780199935383.013.21. ISBN 978-0-19-993538-3. https://www.oxfordhandbooks.com/view/10.1093/oxfordhb/9780199935383.001.0001/oxfordhb-9780199935383-e-21.
- ↑ Gross, T. and Sayama, H. (Eds.). 2009. Adaptive Networks: Theory, Models and Applications. Springer.
- ↑ Holme, P. and Saramäki, J. 2013. Temporal Networks. Springer.
- ↑ Xanthos, Aris, Pante, Isaac, Rochat, Yannick, Grandjean, Martin (2016). Visualising the Dynamics of Character Networks. In Digital Humanities 2016: Jagiellonian University & Pedagogical University, Kraków, pp. 417–419.
- ↑ Barabási, A. L.; Gulbahce, N.; Loscalzo, J. (2011). "Network medicine: a network-based approach to human disease". Nature Reviews Genetics 12 (1): 56–68. doi:10.1038/nrg2918. PMID 21164525.
- ↑ Newman, M., Barabási, A.-L., Watts, D.J. [eds.] (2006) The Structure and Dynamics of Networks. Princeton, N.J.: Princeton University Press.
- ↑ Dorogovtsev, S N; Mendes, J F F (2003). Evolution of Networks: From Biological Nets to the Internet and WWW. New York, NY, USA: Oxford University Press, Inc.. ISBN 978-0198515906.
- ↑ Cotacallapa, M; Hase, M O (2016). "Epidemics in networks: a master equation approach". Journal of Physics A 49 (6): 065001. doi:10.1088/1751-8113/49/6/065001. Bibcode: 2016JPhA...49f5001C.
- ↑ De Domenico, Manlio (March 31, 2022). Multilayer Networks: Analysis and Visualization (1st ed.). Springer.
- ↑ Rossi, Luca; Dickison, Mark E.; Magnani, Matteo (July 18, 2016). Multilayer Social Networks (1st ed.). Cambridge University Press.
- ↑ "Identifying, understanding, and analyzing critical infrastructure interdependencies". IEEE Control Systems Magazine 21 (6): 11–25. December 2001. doi:10.1109/37.969131.
- ↑ Buldyrev, Sergey V. (April 2010). "Catastrophic cascade of failures in interdependent networks". Nature 464 (7291): 1025–1028. doi:10.1038/nature08932. PMID 20393559. Bibcode: 2010Natur.464.1025B.
Further reading
- A First Course in Network Science, F. Menczer, S. Fortunato, C.A. Davis. (Cambridge University Press, 2020). ISBN:9781108471138. GitHub site with tutorials, datasets, and other resources
- "Connected: The Power of Six Degrees," https://web.archive.org/web/20111006191031/http://ivl.slis.indiana.edu/km/movies/2008-talas-connected.mov
- Cohen, R.; Erez, K. (2000). "Resilience of the Internet to random breakdown". Phys. Rev. Lett. 85 (21): 4626–4628. doi:10.1103/physrevlett.85.4626. PMID 11082612. Bibcode: 2000PhRvL..85.4626C. http://havlin.biu.ac.il/Publications.php?keyword=Resilience+of+the+Internet+to+random+breakdown&year=*&match=all.
- Pu, Cun-Lai; Wen-; Pei, Jiang; Michaelson, Andrew (2012). "Robustness analysis of network controllability". Physica A 391 (18): 4420–4425. doi:10.1016/j.physa.2012.04.019. Bibcode: 2012PhyA..391.4420P. http://www.barabasilab.com/pubs/CCNR-ALB_Publications/201204-24_PhysicaA-RobustnessControllability/201204-24_PhysicaA-RobustnessControllability.pdf. Retrieved 2013-09-18.
- S.N. Dorogovtsev and J.F.F. Mendes, Evolution of Networks: From biological networks to the Internet and WWW, Oxford University Press, 2003, ISBN:0-19-851590-1
- Linked: The New Science of Networks, A.-L. Barabási (Perseus Publishing, Cambridge)
- 'Scale-Free Networks, G. Caldarelli (Oxford University Press, Oxford)
- Network Science, Committee on Network Science for Future Army Applications, National Research Council. 2005. The National Academies Press (2005)ISBN:0-309-10026-7
- Network Science Bulletin, USMA (2007) ISBN:978-1-934808-00-9
- The Structure and Dynamics of Networks Mark Newman, Albert-László Barabási, & Duncan J. Watts (The Princeton Press, 2006) ISBN:0-691-11357-2
- Dynamical processes on complex networks, Alain Barrat, Marc Barthelemy, Alessandro Vespignani (Cambridge University Press, 2008) ISBN:978-0-521-87950-7
- Network Science: Theory and Applications, Ted G. Lewis (Wiley, March 11, 2009) ISBN:0-470-33188-7
- Nexus: Small Worlds and the Groundbreaking Theory of Networks, Mark Buchanan (W. W. Norton & Company, June 2003) ISBN:0-393-32442-7
- Six Degrees: The Science of a Connected Age, Duncan J. Watts (W. W. Norton & Company, February 17, 2004) ISBN:0-393-32542-3
 |