Biology:Caspase 3
 Generic protein structure example |
Caspase-3 is a caspase protein that interacts with caspase-8 and caspase-9. It is encoded by the CASP3 gene. CASP3 orthologs[1] have been identified in numerous mammals for which complete genome data are available. Unique orthologs are also present in birds, lizards, lissamphibians, and teleosts.
The CASP3 protein is a member of the cysteine-aspartic acid protease (caspase) family.[2] Sequential activation of caspases plays a central role in the execution-phase of cell apoptosis. Caspases exist as inactive proenzymes that undergo proteolytic processing at conserved aspartic residues to produce two subunits, large and small, that dimerize to form the active enzyme. This protein cleaves and activates caspases 6 and 7; and the protein itself is processed and activated by caspases 8, 9, and 10. It is the predominant caspase involved in the cleavage of amyloid-beta 4A precursor protein, which is associated with neuronal death in Alzheimer's disease.[3] Alternative splicing of this gene results in two transcript variants that encode the same protein.[4]
 |
 |
Caspase-3 shares many of the typical characteristics common to all currently-known caspases. For example, its active site contains a cysteine residue (Cys-163) and histidine residue (His-121) that stabilize the peptide bond cleavage of a protein sequence to the carboxy-terminal side of an aspartic acid when it is part of a particular 4-amino acid sequence.[6][7] This specificity allows caspases to be incredibly selective, with a 20,000-fold preference for aspartic acid over glutamic acid.[8] A key feature of caspases in the cell is that they are present as zymogens, termed procaspases, which are inactive until a biochemical change causes their activation. Each procaspase has an N-terminal large subunit of about 20 kDa followed by a smaller subunit of about 10 kDa, called p20 and p10, respectively.[9]
Substrate specificity
Under normal circumstances, caspases recognize tetra-peptide sequences on their substrates and hydrolyze peptide bonds after aspartic acid residues. Caspase 3 and caspase 7 share similar substrate specificity by recognizing tetra-peptide motif Asp-x-x-Asp.[10] The C-terminal Asp is absolutely required while variations at other three positions can be tolerated.[11] Caspase substrate specificity has been widely used in caspase based inhibitor and drug design.[12]
Structure
Caspase-3, in particular, (also known as CPP32/Yama/apopain)[13][14][15] is formed from a 32 kDa zymogen that is cleaved into 17 kDa and 12 kDa subunits. When the procaspase is cleaved at a particular residue, the active heterotetramer can then be formed by hydrophobic interactions, causing four anti-parallel beta-sheets from p17 and two from p12 to come together to make a heterodimer, which in turn interacts with another heterodimer to form the full 12-stranded beta-sheet structure surrounded by alpha-helices that is unique to caspases.[9][16] When the heterodimers align head-to-tail with each other, an active site is positioned at each end of the molecule formed by residues from both participating subunits, though the necessary Cys-163 and His-121 residues are found on the p17 (larger) subunit.[16]
Mechanism
The catalytic site of caspase-3 involves the thiol group of Cys-163 and the imidazole ring of His-121. His-121 stabilizes the carbonyl group of the key aspartate residue, while Cys-163 attacks to ultimately cleave the peptide bond. Cys-163 and Gly-238 also function to stabilize the tetrahedral transition state of the substrate-enzyme complex through hydrogen bonding.[16] In vitro, caspase-3 has been found to prefer the peptide sequence DEVDG (Asp-Glu-Val-Asp-Gly) with cleavage occurring on the carboxy side of the second aspartic acid residue (between D and G).[8][16][17] Caspase-3 is active over a broad pH range that is slightly higher (more basic) than many of the other executioner caspases. This broad range indicates that caspase-3 will be fully active under normal and apoptotic cell conditions.[18]
Activation
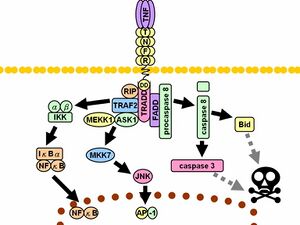
Caspase-3 is activated in the apoptotic cell both by extrinsic (death ligand) and intrinsic (mitochondrial) pathways.[9][19] The zymogen feature of caspase-3 is necessary because if unregulated, caspase activity would kill cells indiscriminately.[20] As an executioner caspase, the caspase-3 zymogen has virtually no activity until it is cleaved by an initiator caspase after apoptotic signaling events have occurred.[21] One such signaling event is the introduction of granzyme B, which can activate initiator caspases, into cells targeted for apoptosis by killer T cells.[22][23] This extrinsic activation then triggers the hallmark caspase cascade characteristic of the apoptotic pathway, in which caspase-3 plays a dominant role.[7] In intrinsic activation, cytochrome c from the mitochondria works in combination with caspase-9, apoptosis-activating factor 1 (Apaf-1), and ATP to process procaspase-3.[17][23][24] These molecules are sufficient to activate caspase-3 in vitro, but other regulatory proteins are necessary in vivo.[24] Mangosteen (Garcinia mangostana) extract has been shown to inhibit the activation of caspase 3 in B-amyloid treated human neuronal cells.[25]
Inhibition
One means of caspase inhibition is through the IAP (inhibitor of apoptosis) protein family, which includes c-IAP1, c-IAP2, XIAP, and ML-IAP.[16] XIAP binds and inhibits initiator caspase-9, which is directly involved in the activation of executioner caspase-3.[24] During the caspase cascade, however, caspase-3 functions to inhibit XIAP activity by cleaving caspase-9 at a specific site, preventing XIAP from being able to bind to inhibit caspase-9 activity.[26]
Interactions
Caspase 3 has been shown to interact with:
Biological function
Caspase-3 has been found to be necessary for normal brain development as well as its typical role in apoptosis, where it is responsible for chromatin condensation and DNA fragmentation.[17] Elevated levels of a fragment of Caspase-3, p17, in the bloodstream is a sign of a recent myocardial infarction.[48] It is now being shown that caspase-3 may play a role in embryonic and hematopoietic stem cell differentiation.[49]
See also
References
- ↑ "OrthoMaM phylogenetic marker: CASP3 coding sequence". http://www.orthomam.univ-montp2.fr/orthomam/data/cds/detailMarkers/ENSG00000164305_CASP3.xml.
- ↑ "Human ICE/CED-3 protease nomenclature". Cell 87 (2): 171. October 1996. doi:10.1016/S0092-8674(00)81334-3. PMID 8861900.
- ↑ "Involvement of caspases in proteolytic cleavage of Alzheimer's amyloid-beta precursor protein and amyloidogenic A beta peptide formation". Cell 97 (3): 395–406. April 1999. doi:10.1016/s0092-8674(00)80748-5. PMID 10319819.
- ↑ "Entrez Gene: CASP3 caspase 3, apoptosis-related cysteine peptidase". https://www.ncbi.nlm.nih.gov/sites/entrez?Db=gene&Cmd=ShowDetailView&TermToSearch=836.
- ↑ "Construction and analysis of a modular model of caspase activation in apoptosis". Theoretical Biology & Medical Modelling 5 (1): 26. 2008. doi:10.1186/1742-4682-5-26. PMID 19077196.
- ↑ "Apoptosis: an overview". British Medical Bulletin 53 (3): 451–65. 1997. doi:10.1093/oxfordjournals.bmb.a011623. PMID 9374030.
- ↑ 7.0 7.1 "Zinc is a potent inhibitor of the apoptotic protease, caspase-3. A novel target for zinc in the inhibition of apoptosis". The Journal of Biological Chemistry 272 (30): 18530–3. July 1997. doi:10.1074/jbc.272.30.18530. PMID 9228015.
- ↑ 8.0 8.1 "Internally quenched fluorescent peptide substrates disclose the subsite preferences of human caspases 1, 3, 6, 7 and 8". The Biochemical Journal 350 (2): 563–8. September 2000. doi:10.1042/0264-6021:3500563. PMID 10947972.
- ↑ 9.0 9.1 9.2 "Caspases: opening the boxes and interpreting the arrows". Cell Death and Differentiation 9 (1): 3–5. January 2002. doi:10.1038/sj.cdd.4400963. PMID 11803369.
- ↑ "Plasticity of S2-S4 specificity pockets of executioner caspase-7 revealed by structural and kinetic analysis". The FEBS Journal 274 (18): 4752–65. September 2007. doi:10.1111/j.1742-4658.2007.05994.x. PMID 17697120.
- ↑ "Structural and kinetic analysis of caspase-3 reveals role for s5 binding site in substrate recognition". Journal of Molecular Biology 360 (3): 654–66. July 2006. doi:10.1016/j.jmb.2006.05.041. PMID 16781734.
- ↑ "Caspases: structure-guided design of drugs to control cell death". Mini Reviews in Medicinal Chemistry 8 (11): 1154–62. October 2008. doi:10.2174/138955708785909899. PMID 18855730.
- ↑ "CPP32, a novel human apoptotic protein with homology to Caenorhabditis elegans cell death protein Ced-3 and mammalian interleukin-1 beta-converting enzyme". The Journal of Biological Chemistry 269 (49): 30761–4. December 1994. doi:10.1016/S0021-9258(18)47344-9. PMID 7983002.
- ↑ "Yama/CPP32 beta, a mammalian homolog of CED-3, is a CrmA-inhibitable protease that cleaves the death substrate poly(ADP-ribose) polymerase". Cell 81 (5): 801–9. June 1995. doi:10.1016/0092-8674(95)90541-3. PMID 7774019.
- ↑ "Identification and inhibition of the ICE/CED-3 protease necessary for mammalian apoptosis". Nature 376 (6535): 37–43. July 1995. doi:10.1038/376037a0. PMID 7596430. Bibcode: 1995Natur.376...37N.
- ↑ 16.0 16.1 16.2 16.3 16.4 "Caspases: pharmacological manipulation of cell death". The Journal of Clinical Investigation 115 (10): 2665–72. October 2005. doi:10.1172/JCI26252. PMID 16200200.
- ↑ 17.0 17.1 17.2 "Emerging roles of caspase-3 in apoptosis". Cell Death and Differentiation 6 (2): 99–104. February 1999. doi:10.1038/sj.cdd.4400476. PMID 10200555.
- ↑ "Biochemical characteristics of caspases-3, -6, -7, and -8". The Journal of Biological Chemistry 272 (41): 25719–23. October 1997. doi:10.1074/jbc.272.41.25719. PMID 9325297.
- ↑ "Apoptosis and cancer: mutations within caspase genes". Journal of Medical Genetics 46 (8): 497–510. August 2009. doi:10.1136/jmg.2009.066944. PMID 19505876.
- ↑ "Mechanisms of caspase activation". Current Opinion in Cell Biology 15 (6): 725–31. December 2003. doi:10.1016/j.ceb.2003.10.009. PMID 14644197.
- ↑ "A constitutively active and uninhibitable caspase-3 zymogen efficiently induces apoptosis". The Biochemical Journal 424 (3): 335–45. December 2009. doi:10.1042/BJ20090825. PMID 19788411.
- ↑ "Apoptosis: live or die--hard work either way!". Hormone and Metabolic Research 33 (9): 511–9. September 2001. doi:10.1055/s-2001-17213. PMID 11561209.
- ↑ 23.0 23.1 "Novel procaspase-3 activating cascade mediated by lysoapoptases and its biological significances in apoptosis". Advances in Enzyme Regulation 41 (1): 237–50. 2001. doi:10.1016/S0065-2571(00)00018-2. PMID 11384748.
- ↑ 24.0 24.1 24.2 "Mitochondrial activation of apoptosis". Cell 116 (2 Suppl): S57–9, 2 p following S59. January 2004. doi:10.1016/S0092-8674(04)00031-5. PMID 15055583.
- ↑ "Protective effect of mangosteen extract against beta-amyloid-induced cytotoxicity, oxidative stress and altered proteome in SK-N-SH cells". Journal of Proteome Research 9 (5): 2076–86. May 2010. doi:10.1021/pr100049v. PMID 20232907. http://ntur.lib.ntu.edu.tw/bitstream/246246/243509/-1/134.pdf.
- ↑ "Caspase 3 attenuates XIAP (X-linked inhibitor of apoptosis protein)-mediated inhibition of caspase 9". The Biochemical Journal 405 (1): 11–9. July 2007. doi:10.1042/BJ20070288. PMID 17437405.
- ↑ "Caspase-2 induces apoptosis by releasing proapoptotic proteins from mitochondria". The Journal of Biological Chemistry 277 (16): 13430–7. April 2002. doi:10.1074/jbc.M108029200. PMID 11832478.
- ↑ "Molecular ordering of the Fas-apoptotic pathway: the Fas/APO-1 protease Mch5 is a CrmA-inhibitable protease that activates multiple Ced-3/ICE-like cysteine proteases". Proceedings of the National Academy of Sciences of the United States of America 93 (25): 14486–91. December 1996. doi:10.1073/pnas.93.25.14486. PMID 8962078. Bibcode: 1996PNAS...9314486S.
- ↑ Selvakumar, P.; Sharma, RK. (May 2007). "Role of calpain and caspase system in the regulation of N-myristoyltransferase in human colon cancer (Review).". Int J Mol Med 19 (5): 823–7. doi:10.3892/ijmm.19.5.823. PMID 17390089.
- ↑ "Casper is a FADD- and caspase-related inducer of apoptosis". Immunity 6 (6): 751–63. June 1997. doi:10.1016/S1074-7613(00)80450-1. PMID 9208847.
- ↑ "MRIT, a novel death-effector domain-containing protein, interacts with caspases and BclXL and initiates cell death". Proceedings of the National Academy of Sciences of the United States of America 94 (21): 11333–8. October 1997. doi:10.1073/pnas.94.21.11333. PMID 9326610. Bibcode: 1997PNAS...9411333H.
- ↑ "The dependence receptor DCC (deleted in colorectal cancer) defines an alternative mechanism for caspase activation". Proceedings of the National Academy of Sciences of the United States of America 98 (6): 3416–21. March 2001. doi:10.1073/pnas.051378298. PMID 11248093. Bibcode: 2001PNAS...98.3416F.
- ↑ "Presence of a pre-apoptotic complex of pro-caspase-3, Hsp60 and Hsp10 in the mitochondrial fraction of jurkat cells". The EMBO Journal 18 (8): 2040–8. April 1999. doi:10.1093/emboj/18.8.2040. PMID 10205158.
- ↑ "Hsp60 accelerates the maturation of pro-caspase-3 by upstream activator proteases during apoptosis". The EMBO Journal 18 (8): 2049–56. April 1999. doi:10.1093/emboj/18.8.2049. PMID 10205159.
- ↑ "Protein kinase CK2 inhibitor 4,5,6,7-tetrabromobenzotriazole (TBB) induces apoptosis and caspase-dependent degradation of haematopoietic lineage cell-specific protein 1 (HS1) in Jurkat cells". The Biochemical Journal 364 (Pt 1): 41–7. May 2002. doi:10.1042/bj3640041. PMID 11988074.
- ↑ "Caspase-mediated cleavage of actin-binding and SH3-domain-containing proteins cortactin, HS1, and HIP-55 during apoptosis". Biochemical and Biophysical Research Communications 288 (4): 981–9. November 2001. doi:10.1006/bbrc.2001.5862. PMID 11689006.
- ↑ "IAP-family protein survivin inhibits caspase activity and apoptosis induced by Fas (CD95), Bax, caspases, and anticancer drugs". Cancer Research 58 (23): 5315–20. December 1998. PMID 9850056.
- ↑ "An anti-apoptotic protein human survivin is a direct inhibitor of caspase-3 and -7". Biochemistry 40 (4): 1117–23. January 2001. doi:10.1021/bi001603q. PMID 11170436.
- ↑ "Caspase-mediated cleavage of TRAF3 in FasL-stimulated Jurkat-T cells". Journal of Leukocyte Biology 69 (3): 490–6. March 2001. doi:10.1189/jlb.69.3.490. PMID 11261798.
- ↑ "TRAF1 is a substrate of caspases activated during tumor necrosis factor receptor-alpha-induced apoptosis". The Journal of Biological Chemistry 276 (11): 8087–93. March 2001. doi:10.1074/jbc.M009450200. PMID 11098060.
- ↑ "Ubiquitin-protein ligase activity of X-linked inhibitor of apoptosis protein promotes proteasomal degradation of caspase-3 and enhances its anti-apoptotic effect in Fas-induced cell death". Proceedings of the National Academy of Sciences of the United States of America 98 (15): 8662–7. July 2001. doi:10.1073/pnas.161506698. PMID 11447297. Bibcode: 2001PNAS...98.8662S.
- ↑ "The anti-apoptotic activity of XIAP is retained upon mutation of both the caspase 3- and caspase 9-interacting sites". The Journal of Cell Biology 157 (1): 115–24. April 2002. doi:10.1083/jcb.200108085. PMID 11927604.
- ↑ "Structural basis for the inhibition of caspase-3 by XIAP". Cell 104 (5): 791–800. March 2001. doi:10.1016/S0092-8674(01)00274-4. PMID 11257232.
- ↑ "The c-IAP-1 and c-IAP-2 proteins are direct inhibitors of specific caspases". The EMBO Journal 16 (23): 6914–25. December 1997. doi:10.1093/emboj/16.23.6914. PMID 9384571.
- ↑ "X-linked IAP is a direct inhibitor of cell-death proteases". Nature 388 (6639): 300–4. July 1997. doi:10.1038/40901. PMID 9230442. Bibcode: 1997Natur.388..300D.
- ↑ "X-linked inhibitor of apoptosis protein (XIAP) inhibits caspase-3 and -7 in distinct modes". The Journal of Biological Chemistry 276 (29): 27058–63. July 2001. doi:10.1074/jbc.M102415200. PMID 11359776.
- ↑ "Identification of NRF2, a member of the NF-E2 family of transcription factors, as a substrate for caspase-3(-like) proteases". Cell Death and Differentiation 6 (9): 865–72. September 1999. doi:10.1038/sj.cdd.4400566. PMID 10510468.
- ↑ "Serum caspase-3 p17 fragment is elevated in patients with ST-segment elevation myocardial infarction: a novel observation". Journal of the American College of Cardiology 57 (2): 220–1. January 2011. doi:10.1016/j.jacc.2010.08.628. PMID 21211695.
- ↑ "Rehabilitation of a contract killer: caspase-3 directs stem cell differentiation". Cell Stem Cell 2 (6): 515–6. June 2008. doi:10.1016/j.stem.2008.05.013. PMID 18522841.
Further reading
- "Caspases: the executioners of apoptosis". The Biochemical Journal 326 (Pt 1): 1–16. August 1997. doi:10.1042/bj3260001. PMID 9337844.
- Cytostatic p21 G protein-activated protein kinase gamma-PAK. Vitamins & Hormones. 62. 2001. pp. 167–98. doi:10.1016/S0083-6729(01)62004-1. ISBN 9780127098623.
- "Structure and function of HIV-1 auxiliary regulatory protein Vpr: novel clues to drug design". Current Drug Targets. Immune, Endocrine and Metabolic Disorders 4 (4): 265–75. December 2004. doi:10.2174/1568008043339668. PMID 15578977.
- "The Vpr protein from HIV-1: distinct roles along the viral life cycle". Retrovirology 2 (1): 11. 2006. doi:10.1186/1742-4690-2-11. PMID 15725353.
- "Reversible glutathiolation of caspase-3 by glutaredoxin as a novel redox signaling mechanism in tumor necrosis factor-alpha-induced cell death". Circulation Research 100 (2): 152–4. February 2007. doi:10.1161/01.RES.0000258171.08020.72. PMID 17272816.
External links
- The MEROPS online database for peptidases and their inhibitors: C14.003
- Apoptosis & Caspase 3 – The Proteolysis Map-animation
 |