Biology:Caspase-9
 Generic protein structure example |
Caspase-9 is an enzyme that in humans is encoded by the CASP9 gene. It is an initiator caspase,[1] critical to the apoptotic pathway found in many tissues.[2] Caspase-9 homologs have been identified in all mammals for which they are known to exist, such as Mus musculus and Pan troglodytes.[3]
Caspase-9 belongs to a family of caspases, cysteine-aspartic proteases involved in apoptosis and cytokine signalling.[4] Apoptotic signals cause the release of cytochrome c from mitochondria and activation of apaf-1 (apoptosome), which then cleaves the pro-enzyme of caspase-9 into the active dimer form.[2] Regulation of this enzyme occurs through phosphorylation by an allosteric inhibitor, inhibiting dimerization and inducing a conformational change.[4]
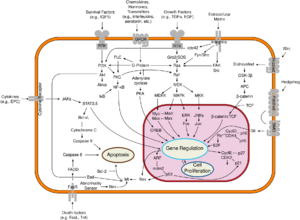
Correct caspase-9 function is required for apoptosis, leading to the normal development of the central nervous system.[4] Caspase-9 has multiple additional cellular functions that are independent of its role in apoptosis. Nonapoptotic roles of caspase-9 include regulation of necroptosis, cellular differentiation, innate immune response, sensory neuron maturation, mitochondrial homeostasis, corticospinal circuit organization, and ischemic vascular injury. [5] Without correct function, abnormal tissue development can occur leading to abnormal function, diseases and premature death. [4] Caspase-9 loss-of-function mutations have been associated with immunodeficiency/lymphoproliferation, neural tube defects, and Li-Fraumeni-like syndrome. Increased caspase-9 activity is implicated in the progression of amyotrophic lateral sclerosis, retinal detachment, and slow-channel syndrome, as well as various other neurological, autoimmune, and cardiovascular disorders. [5]
Different protein isoforms of caspase-9 are produced due to alternative splicing.[6]
Structure
Similar to other caspases, caspase-9 has three domains: N-terminal pro-domain, large subunit, and a small subunit.[4] The N-terminal pro-domain is also called the long pro-domain and this contains the caspase activation domain (CARD) motif.[7] The pro-domain is linked to the catalytic domain by a linker loop.[8]
The caspase-9 monomer consists of one large and one small subunit, both comprising the catalytic domain.[9] Differing from the normally conserved active site motif QACRG in other caspases, caspase-9 has the motif QACGG.[10][8]
When dimerized, caspase-9 has two different active site conformations within each dimer.[9] One site closely resembles the catalytic site of other caspases, whereas the second has no 'activation loop', disrupting the catalytic machinery in that particular active site.[9] Surface loops around the active site are short, giving rise to broad substrate specificity as the substrate-binding cleft is more open.[11] Within caspase-9's active site, in order for catalytic activity to occur there has to be specific amino acids in the right position. Amino acid Asp at position P1 is essential, with a preference for amino acid His at position P2.[12]
Localization
Within the cell, caspase-9 in humans is found in the mitochondria, cytosol, and nucleus.[13]
Protein expression
Caspase-9 in humans is expressed in fetus and adult tissues.[10][8] Tissue expression of caspase-9 is ubiquitous with the highest expression in the brain and heart, specifically at the developmental stage of an adult in the heart's muscle cells.[14] The liver, pancreas, and skeletal muscle express this enzyme at a moderate level, and all other tissues express caspase-9 at low levels.[14]
Mechanism
Active caspase-9 works as an initiating caspase by cleaving, thus activating downstream executioner caspases, initiating apoptosis.[15] Once activated, caspase-9 goes on to cleave caspase-3, -6, and -7, initiating the caspase cascade as they cleave several other cellular targets.[4]
When caspase-9 is inactive, it exists in the cytosol as a zymogen, in its monomer form.[9][16] It is then recruited and activated by the CARDs in apaf-1, recognizing the CARDs in caspase-9.[17]
Processing
Before activation can occur, caspase-9 has to be processed.[18] Initially, caspase-9 is made as an inactive single-chain zymogen.[18] Processing occurs when the apoptosome binds to pro-caspase-9 as apaf-1 assists in the autoproteolytic processing of the zymogen.[18] The processed caspase-9 stays bound to the apoptosome complex, forming a holoenzyme.[19]
Activation
Activation occurs when caspase-9 dimerizes, and there are two different ways for which this can occur:
- Caspase-9 is auto-activated when it binds to apaf-1(apoptosome), as apaf-1 oligomerizes the precursor molecules of pro-caspase-9.[13]
- Previously activated caspases can cleave caspase-9, causing its dimerization.[20]
Catalytic activity
Caspase-9 has a preferred cleavage sequence of Leu-Gly-His-Asp-(cut)-X.[12]
Regulation
Negative regulation of caspase-9 occurs through phosphorylation.[4] This is done by a serine-threonine kinase, Akt, on serine-196 which inhibits the activation and protease activity of caspase-9, suppressing caspase-9 and further activation of apoptosis.[21] Akt acts as an allosteric inhibitor of caspase-9 because the site of phosphorylation of serine-196 is far from the catalytic site.[21] The inhibitor affects the dimerization of caspase-9 and causes a conformational change that affects the substrate-binding cleft of caspase-9.[21]
Akt can act on both processed and unprocessed caspase-9 in-vitro, where phosphorylation on processed caspase-9 occurs on the large subunit.[22]
Deficiencies and mutations
A deficiency in caspase-9 largely affects the brain and its development.[23] The effects of having a mutation or deficiency in this caspase compared to others is detrimental.[23] The initiating role caspase-9 plays in apoptosis is the cause for the severe effects seen in those with an atypical caspase-9.
Mice with insufficient caspase-9 have a main phenotype of an affected or abnormal brain.[4] Larger brains due to a decrease in apoptosis, resulting in an increase of extra neurons is an example of a phenotype seen in caspase-9 deficient mice.[24] Those homozygous for no caspase-9 die perinatally as a result of an abnormally developed cerebrum.[4]
In humans, expression of caspase-9 varies from tissue to tissue, and the different levels have a physiological role.[24] Low amounts of caspase-9 leads to cancer and neurodegenerative diseases like Alzheimer's disease.[24] Further alterations at single-nucleotide polymorphism (SNP) levels and whole gene levels of caspase-9 can cause germ-line mutations linked to non-Hodgkin's lymphoma.[25] Certain polymorphisms in the promoter of caspase-9 enhances the rate at which caspase-9 is expressed, and this can increase a person's risk of lung cancer.[26]
Clinical significance
The effects of abnormal caspase-9 levels or function impacts the clinical world. The impact caspase-9 has on the brain can lead to future work in inhibition through targeted therapy, specifically with diseases associated with the brain as this enzyme may take part in the developmental pathways of neuronal disorders.[4]
The introduction of caspases may also have medical benefits.[15] In the context of graft versus host disease, caspase-9 can be introduced as an inducible switch.[27] In the presence of a small molecule, it will dimerize and trigger apoptosis, eliminating lymphocytes.[27]
iCasp9
iCasp9 (inducible caspase-9) is a type of control system for chimeric antigen receptor T cells (CAR T cells). CAR T cells are genetically modified T cells that exhibit cytotoxicity to tumor cells. Evidence shows that CAR T cells are effective in treating B-cell malignancies. However, as CAR T cells introduce toxicity, user control of the cells and their targets is critical.[28] One of the various ways to exert control over CAR T cell is through drug-controlled synthetic systems. iCasp9 was created by modifying caspase-9 and fusing it with the FK506 binding protein.[28] iCasp9 can be added to the CAR T cells as an inducible suicide gene.[29]
If therapy with CAR T cells results in severe side effects, iCasp9 can be used to halt treatment. Administering a small-molecule drug such as rapamycin causes the drug to bind to the FK506 domain.[29] This, in turn, induces expression of caspase-9, which triggers cell death of the CAR T cells.[29]
Alternative transcripts
Through alternative splicing, four difference caspase-9 variants are produced.
Caspase-9α (9L)
This variant is used as the reference sequence, and it has full cysteine protease activity.[7][30]
Caspase-9β (9S)
Isoform 2 doesn't include exons 3, 4, 5, and 6; it is missing amino acids 140-289.[7][30] Caspase-9S doesn't have central catalytic domain, therefore it functions as an inhibitor of caspase-9α by attaching to the apoptosome, suppressing the caspase enzyme cascade and apoptosis.[7][31] Caspase-9β is referred to as the endogenous dominant-negative isoform.
Caspase-9γ
This variant is missing amino acids 155-416, and for amino acids 152-154, the sequence AYI is changed to TVL.[30]
Isoform 4
In comparison with the reference sequence, it is missing amino acids 1-83.[30]
Interactions
Caspase-9 has been shown to interact with:
See also
References
- ↑ Caspase 9
- ↑ 2.0 2.1 2.2 "Cytochrome c and dATP-dependent formation of Apaf-1/caspase-9 complex initiates an apoptotic protease cascade". Cell 91 (4): 479–89. November 1997. doi:10.1016/s0092-8674(00)80434-1. PMID 9390557.
- ↑ "HomoloGene - NCBI". https://www.ncbi.nlm.nih.gov/homologene/31024.
- ↑ 4.00 4.01 4.02 4.03 4.04 4.05 4.06 4.07 4.08 4.09 "Caspase-9". The International Journal of Biochemistry & Cell Biology 32 (2): 121–4. 2000. doi:10.1016/s1357-2725(99)00024-2. PMID 10687948.
- ↑ 5.0 5.1 "Caspase-9: A Multimodal Therapeutic Target With Diverse Cellular Expression in Human Disease". Frontiers in Pharmacology 12: 701301. 2021. doi:10.3389/fphar.2021.701301. PMID 34305609.
- ↑ "CASP9 caspase 9 [Homo sapiens (human) - Gene - NCBI"]. https://www.ncbi.nlm.nih.gov/gene/842.
- ↑ 7.0 7.1 7.2 7.3 "Caspase-9: structure, mechanisms and clinical application". Oncotarget 8 (14): 23996–24008. April 2017. doi:10.18632/oncotarget.15098. PMID 28177918.
- ↑ 8.0 8.1 8.2 "The Ced-3/interleukin 1beta converting enzyme-like homolog Mch6 and the lamin-cleaving enzyme Mch2alpha are substrates for the apoptotic mediator CPP32". The Journal of Biological Chemistry 271 (43): 27099–106. October 1996. doi:10.1074/jbc.271.43.27099. PMID 8900201.
- ↑ 9.0 9.1 9.2 9.3 "Dimer formation drives the activation of the cell death protease caspase 9". Proceedings of the National Academy of Sciences of the United States of America 98 (25): 14250–5. December 2001. doi:10.1073/pnas.231465798. PMID 11734640. Bibcode: 2001PNAS...9814250R.
- ↑ 10.0 10.1 "ICE-LAP6, a novel member of the ICE/Ced-3 gene family, is activated by the cytotoxic T cell protease granzyme B". The Journal of Biological Chemistry 271 (28): 16720–4. July 1996. doi:10.1074/jbc.271.28.16720. PMID 8663294.
- ↑ "A combinatorial approach defines specificities of members of the caspase family and granzyme B. Functional relationships established for key mediators of apoptosis". The Journal of Biological Chemistry 272 (29): 17907–11. July 1997. doi:10.1074/jbc.272.29.17907. PMID 9218414.
- ↑ 12.0 12.1 "The E. coli effector protein NleF is a caspase inhibitor". PLOS ONE 8 (3): e58937. 2013-03-14. doi:10.1371/journal.pone.0058937. PMID 23516580. Bibcode: 2013PLoSO...858937B.
- ↑ 13.0 13.1 "Caspases: their intracellular localization and translocation during apoptosis". Cell Death and Differentiation 6 (7): 644–51. July 1999. doi:10.1038/sj.cdd.4400536. PMID 10453075.
- ↑ 14.0 14.1 "Overexpression of HAX-1 protects cardiac myocytes from apoptosis through caspase-9 inhibition". Circulation Research 99 (4): 415–23. August 2006. doi:10.1161/01.RES.0000237387.05259.a5. PMID 16857965.
- ↑ 15.0 15.1 "Caspase functions in cell death and disease". Cold Spring Harbor Perspectives in Biology 5 (4): a008656. April 2013. doi:10.1101/cshperspect.a008656. PMID 23545416.
- ↑ "Caspase functions in cell death and disease". Cold Spring Harbor Perspectives in Biology 5 (4): a008656. April 2013. doi:10.1101/cshperspect.a008656. PMID 23545416.
- ↑ "Three-dimensional structure of the apoptosome: implications for assembly, procaspase-9 binding, and activation". Molecular Cell 9 (2): 423–32. 2002. doi:10.1016/s1097-2765(02)00442-2. PMID 11864614.
- ↑ 18.0 18.1 18.2 "Proteolytic processing of the caspase-9 zymogen is required for apoptosome-mediated activation of caspase-9". The Journal of Biological Chemistry 288 (21): 15142–7. May 2013. doi:10.1074/jbc.M112.441568. PMID 23572523.
- ↑ "Molecular cell death platforms and assemblies". Current Opinion in Cell Biology 22 (6): 828–36. December 2010. doi:10.1016/j.ceb.2010.08.004. PMID 20817427.
- ↑ "Overexpression of caspase-9 triggers its activation and apoptosis in vitro". Croatian Medical Journal 47 (6): 832–40. December 2006. PMID 17167855.
- ↑ 21.0 21.1 21.2 "Regulation of cell death protease caspase-9 by phosphorylation". Science 282 (5392): 1318–21. November 1998. doi:10.1126/science.282.5392.1318. PMID 9812896. Bibcode: 1998Sci...282.1318C.
- ↑ "Regulation of cell death protease caspase-9 by phosphorylation". Science 282 (5392): 1318–21. November 1998. doi:10.1126/science.282.5392.1318. PMID 9812896. Bibcode: 1998Sci...282.1318C.
- ↑ 23.0 23.1 "Cell death in brain development and degeneration: control of caspase expression may be key!". Molecular Neurobiology 37 (1): 1–6. February 2008. doi:10.1007/s12035-008-8021-4. PMID 18449809.
- ↑ 24.0 24.1 24.2 "Differential requirement for caspase 9 in apoptotic pathways in vivo". Cell 94 (3): 339–52. 1998. doi:10.1016/s0092-8674(00)81477-4. PMID 9708736.
- ↑ "Germline variation in apoptosis pathway genes and risk of non-Hodgkin's lymphoma". Cancer Epidemiology, Biomarkers & Prevention 19 (11): 2847–58. November 2010. doi:10.1158/1055-9965.EPI-10-0581. PMID 20855536.
- ↑ "Caspase 9 promoter polymorphisms and risk of primary lung cancer". Human Molecular Genetics 15 (12): 1963–71. June 2006. doi:10.1093/hmg/ddl119. PMID 16687442.
- ↑ 27.0 27.1 "An inducible caspase 9 safety switch for T-cell therapy". Blood 105 (11): 4247–54. June 2005. doi:10.1182/blood-2004-11-4564. PMID 15728125.
- ↑ 28.0 28.1 "Engineering T Cells to Treat Cancer: The Convergence of Immuno-Oncology and Synthetic Biology". Annual Review of Cancer Biology 4: 121–139. 2020. doi:10.1146/annurev-cancerbio-030419-033657.
- ↑ 29.0 29.1 29.2 "Definition of autologous iCASP9-CD19-expressing T lymphocytes". https://www.cancer.gov/publications/dictionaries/cancer-drug/def/791844.
- ↑ 30.0 30.1 30.2 30.3 "CASP9 - Caspase-9 precursor - Homo sapiens (Human) - CASP9 gene & protein". https://www.uniprot.org/uniprot/P55211#ptm_processing.
- ↑ "hnRNP U enhances caspase-9 splicing and is modulated by AKT-dependent phosphorylation of hnRNP L". The Journal of Biological Chemistry 288 (12): 8575–84. March 2013. doi:10.1074/jbc.M112.443333. PMID 23396972.
- ↑ 32.0 32.1 "A novel enhancer of the Apaf1 apoptosome involved in cytochrome c-dependent caspase activation and apoptosis". The Journal of Biological Chemistry 276 (12): 9239–45. March 2001. doi:10.1074/jbc.M006309200. PMID 11113115.
- ↑ "Induced inhibition of ischemic/hypoxic injury by APIP, a novel Apaf-1-interacting protein". The Journal of Biological Chemistry 279 (38): 39942–50. September 2004. doi:10.1074/jbc.M405747200. PMID 15262985.
- ↑ "Bcl-XL interacts with Apaf-1 and inhibits Apaf-1-dependent caspase-9 activation". Proceedings of the National Academy of Sciences of the United States of America 95 (8): 4386–91. April 1998. doi:10.1073/pnas.95.8.4386. PMID 9539746. Bibcode: 1998PNAS...95.4386H.
- ↑ "Caspase-9, Bcl-XL, and Apaf-1 form a ternary complex". The Journal of Biological Chemistry 273 (10): 5841–5. March 1998. doi:10.1074/jbc.273.10.5841. PMID 9488720.
- ↑ 36.0 36.1 36.2 "IAPs block apoptotic events induced by caspase-8 and cytochrome c by direct inhibition of distinct caspases". The EMBO Journal 17 (8): 2215–23. April 1998. doi:10.1093/emboj/17.8.2215. PMID 9545235.
- ↑ "Caspase-2 induces apoptosis by releasing proapoptotic proteins from mitochondria". The Journal of Biological Chemistry 277 (16): 13430–7. April 2002. doi:10.1074/jbc.M108029200. PMID 11832478.
- ↑ "Molecular ordering of the Fas-apoptotic pathway: the Fas/APO-1 protease Mch5 is a CrmA-inhibitable protease that activates multiple Ced-3/ICE-like cysteine proteases". Proceedings of the National Academy of Sciences of the United States of America 93 (25): 14486–91. December 1996. doi:10.1073/pnas.93.25.14486. PMID 8962078. Bibcode: 1996PNAS...9314486S.
- ↑ "Molecular cloning and characterization of DEFCAP-L and -S, two isoforms of a novel member of the mammalian Ced-4 family of apoptosis proteins". The Journal of Biological Chemistry 276 (12): 9230–8. March 2001. doi:10.1074/jbc.M009853200. PMID 11076957.
- ↑ "Towards a proteome-scale map of the human protein-protein interaction network". Nature 437 (7062): 1173–8. October 2005. doi:10.1038/nature04209. PMID 16189514. Bibcode: 2005Natur.437.1173R.
- ↑ "Neuronal apoptosis-inhibitory protein does not interact with Smac and requires ATP to bind caspase-9". The Journal of Biological Chemistry 279 (39): 40622–8. September 2004. doi:10.1074/jbc.M405963200. PMID 15280366.
- ↑ "Molecular cloning of ILP-2, a novel member of the inhibitor of apoptosis protein family". Molecular and Cellular Biology 21 (13): 4292–301. July 2001. doi:10.1128/MCB.21.13.4292-4301.2001. PMID 11390657.
Further reading
- "Caspases: the executioners of apoptosis". The Biochemical Journal 326 (Pt 1): 1–16. August 1997. doi:10.1042/bj3260001. PMID 9337844.
- "IAP family proteins--suppressors of apoptosis". Genes & Development 13 (3): 239–52. February 1999. doi:10.1101/gad.13.3.239. PMID 9990849.
- "Structure and function of HIV-1 auxiliary regulatory protein Vpr: novel clues to drug design". Current Drug Targets. Immune, Endocrine and Metabolic Disorders 4 (4): 265–75. December 2004. doi:10.2174/1568008043339668. PMID 15578977.
- "The Vpr protein from HIV-1: distinct roles along the viral life cycle". Retrovirology 2: 11. February 2005. doi:10.1186/1742-4690-2-11. PMID 15725353.
- "Role of HIV Vpr as a regulator of apoptosis and an effector on bystander cells". Molecules and Cells 21 (1): 7–20. February 2006. PMID 16511342.
- "Reproducibility of response to a questionnaire on symptoms of masticatory dysfunction". Community Dentistry and Oral Epidemiology 4 (5): 205–9. September 1976. doi:10.1111/j.1600-0528.1976.tb00985.x. PMID 1067155.
- "CPP32, a novel human apoptotic protein with homology to Caenorhabditis elegans cell death protein Ced-3 and mammalian interleukin-1 beta-converting enzyme". The Journal of Biological Chemistry 269 (49): 30761–4. December 1994. doi:10.1016/S0021-9258(18)47344-9. PMID 7983002.
- "ICE-LAP6, a novel member of the ICE/Ced-3 gene family, is activated by the cytotoxic T cell protease granzyme B". The Journal of Biological Chemistry 271 (28): 16720–4. July 1996. doi:10.1074/jbc.271.28.16720. PMID 8663294.
- "The Ced-3/interleukin 1beta converting enzyme-like homolog Mch6 and the lamin-cleaving enzyme Mch2alpha are substrates for the apoptotic mediator CPP32". The Journal of Biological Chemistry 271 (43): 27099–106. October 1996. doi:10.1074/jbc.271.43.27099. PMID 8900201.
- "Molecular ordering of the Fas-apoptotic pathway: the Fas/APO-1 protease Mch5 is a CrmA-inhibitable protease that activates multiple Ced-3/ICE-like cysteine proteases". Proceedings of the National Academy of Sciences of the United States of America 93 (25): 14486–91. December 1996. doi:10.1073/pnas.93.25.14486. PMID 8962078. Bibcode: 1996PNAS...9314486S.
- "Caspase-3-generated fragment of gelsolin: effector of morphological change in apoptosis". Science 278 (5336): 294–8. October 1997. doi:10.1126/science.278.5336.294. PMID 9323209.
- "Cytochrome c and dATP-dependent formation of Apaf-1/caspase-9 complex initiates an apoptotic protease cascade". Cell 91 (4): 479–89. November 1997. doi:10.1016/S0092-8674(00)80434-1. PMID 9390557.
- "Caspase-9, Bcl-XL, and Apaf-1 form a ternary complex". The Journal of Biological Chemistry 273 (10): 5841–5. March 1998. doi:10.1074/jbc.273.10.5841. PMID 9488720.
- "Bcl-XL interacts with Apaf-1 and inhibits Apaf-1-dependent caspase-9 activation". Proceedings of the National Academy of Sciences of the United States of America 95 (8): 4386–91. April 1998. doi:10.1073/pnas.95.8.4386. PMID 9539746. Bibcode: 1998PNAS...95.4386H.
- "IAPs block apoptotic events induced by caspase-8 and cytochrome c by direct inhibition of distinct caspases". The EMBO Journal 17 (8): 2215–23. April 1998. doi:10.1093/emboj/17.8.2215. PMID 9545235.
- "Autoactivation of procaspase-9 by Apaf-1-mediated oligomerization". Molecular Cell 1 (7): 949–57. June 1998. doi:10.1016/S1097-2765(00)80095-7. PMID 9651578.
- "A cloning method for caspase substrates that uses the yeast two-hybrid system: cloning of the antiapoptotic gene gelsolin". Proceedings of the National Academy of Sciences of the United States of America 95 (15): 8532–7. July 1998. doi:10.1073/pnas.95.15.8532. PMID 9671712. Bibcode: 1998PNAS...95.8532K.
- "Regulation of cell death protease caspase-9 by phosphorylation". Science 282 (5392): 1318–21. November 1998. doi:10.1126/science.282.5392.1318. PMID 9812896. Bibcode: 1998Sci...282.1318C.
- "WD-40 repeat region regulates Apaf-1 self-association and procaspase-9 activation". The Journal of Biological Chemistry 273 (50): 33489–94. December 1998. doi:10.1074/jbc.273.50.33489. PMID 9837928.
- "The Bax subfamily of Bcl2-related proteins is essential for apoptotic signal transduction by c-Jun NH(2)-terminal kinase". Molecular and Cellular Biology 22 (13): 4929–42. July 2002. doi:10.1128/MCB.22.13.4929-4942.2002. PMID 12052897.
- "Mammalian caspases: structure, activation, substrates, and functions during apoptosis". Annual Review of Biochemistry 68: 383–424. 1999. doi:10.1146/annurev.biochem.68.1.383. PMID 10872455.
External links
 |