Biology:Haplogroup X (mtDNA)
| Haplogroup X | |
|---|---|
 | |
| Possible time of origin | ca. 45,000–20,000 years ago[1] |
| Possible place of origin | Near East[2] |
| Ancestor | N |
| Descendants | X1, X2 |
| Defining mutations | 73, 7028, 11719, 12705, 14766, 16189, 16223, 16278[3] |
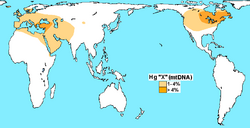
Haplogroup X is a human mitochondrial DNA (mtDNA) haplogroup. It is found in North America, Europe, Western Asia, North Africa, and the Horn of Africa.
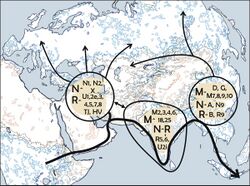
Haplogroup X diverged from haplogroup N, roughly 30,000 years ago (just prior to or during the Last Glacial Maximum). It is in turn ancestral to subclades X2 and X1, which arose c.16-21 thousand and c.14-24 thousand years ago, respectively.[1]
Distribution
Haplogroup X is found in approximately 2% of native Europeans,[4] and 13% of all native North Americans.[5][6] Additionally, the Haplogroup is present in around 3% of Assyrians, with high concentrations in Erzurum, Turkey as well.[citation needed] Notably, the haplogroup is especially common, at 14.3%, among the natives of Bahariya Oasis (Western Desert, Egypt.[7] The X1 subclade is much less frequent, and is largely restricted to North Africa, the Horn of Africa and the Near East.
Subclade X2 appears to have undergone extensive population expansion and dispersal around or soon after the Last Glacial Maximum, roughly 20,000 years ago. It is more strongly represented in the Near East, the Caucasus, and southern Europe, and somewhat less strongly present in the rest of Europe. The highest concentrations are found in the Ojibwe (25%), Sioux (15%), Nuu-Chah-Nulth (12%), Georgia (8%), Orkney (7%), and amongst the Druze Assyrian community in Israel (27%). Subclades of X2 are not present in South Americans Amerindian populations.[8] The oldest known human associated with X2 is Kennewick Man, whose c. 9000-year old remains were discovered in Washington (state) .
Archaeogenetics
Haplogroup X has been found in various other bone specimens that were analysed for ancient DNA, including specimens associated with the Alföld Linear Pottery (X2b-T226C, Garadna-Elkerülő út site 2, 1/1 or 100%), Linearbandkeramik (X2d1, Halberstadt-Sonntagsfeld, 1/22 or ~5%), and Iberia Chalcolithic (X2b, La Chabola de la Hechicera, 1/3 or 33%; X2b, El Sotillo, 1/3 or 33%; X2b, El Mirador Cave, 1/12 or ~8%) cultures.[9] Abel-beth-maachah 2201 was a man who lived between 1014 and 836 BC during the Levant Iron Age and was found in the region now known as Abel Beth Maacah, Metula, Israel. He was associated with the Galilean cultural group. His direct maternal line belonged to mtDNA haplogroup X2b. Haplogroup X has been found in ancient Assyria and ancient Egyptian mummies excavated at the Abusir el-Meleq archaeological site in Middle Egypt, which date from the late New Kingdom and Roman periods.[10] Fossils excavated at the Late Neolithic site of Kelif el Boroud in Morocco, which have been dated to around 5,000 years old, have also been found to carry the X2 subclade.[11]
Druze
In Eurasia, the greatest frequency of haplogroup X is observed in the Druze, a minority population in Israel, Jordan, Lebanon, and Syria, as much in X1 (16%) as in X2 (11%).[12] The Druze also have much diversity of X lineages. This pattern of heterogeneous parental origins is consistent with Druze oral tradition. The Galilee Druze represent a population isolate, so their combination of a high frequency and diversity of X signifies a phylogenetic refugium, providing a sample snapshot of the genetic landscape of the Near East prior to the modern age.[13]
North America
Haplogroup X is also one of the five haplogroups found in the indigenous peoples of the Americas.[14] (namely, X2a subclade).
Although it occurs only at a frequency of about 3% for the total current indigenous population of the Americas, it is a bigger haplogroup in northern North America, where among the Algonquian peoples it comprises up to 25% of mtDNA types.[15][16] It is also present in lesser percentages to the west and south of this area—among the Sioux (15%), the Nuu-chah-nulth (11%–13%), the Navajo (7%), and the Yakama (5%).[17][18] In Latin America, Haplotype X6 was present in the Tarahumara 1.8% (1/53) and Huichol 20% (3/15)[19] X6 and X7 was also found in 12% in Yanomani people.[20]
Unlike the four main Native American mtDNA haplogroups (A, B, C, D), X is not strongly associated with East Asia. The main occurrence of X in Asia discovered so far is in the Altai people in Siberia.[21]
One theory of how the X Haplogroup ended up in North America is that the people carrying it migrated from central Asia along with haplogroups A, B, C, and D, from an ancestor from the Altai Region of Central Asia.[12] Two sequences of haplogroup X2 were sampled further east of Altai among the Evenks of Central Siberia.[12] These two sequences belong to X2* and X2b. It is uncertain if they represent a remnant of the migration of X2 through Siberia or a more recent input.[12]
This relative absence of haplogroup X2 in Asia is one of the major factors used to support the Solutrean hypothesis during the early 2000s. The Solutrean hypothesis postulates that haplogroup X reached North America with a wave of European migration emerging from the Solutrean culture, a stone-age culture in south-western France and in Spain , by boat around the southern edge of the Arctic ice pack roughly 20,000 years ago.[22][23] Since the later 2000s and during the 2010s, evidence has turned against the Solutrean hypothesis, as no presence of mt-DNA ancestral to X2a has been found in Europe or the Near East. New World lineages X2a and X2g are not derived from the Old World lineages X2b, X2c, X2d, X2e, and X2f, indicating an early origin of the New World lineages "likely at the very beginning of their expansion and spread from the Near East".[12] A 2008 study came to the conclusion that the presence of haplogroup X in the Americas does not support migration from Solutrean-period Europe.[17] The lineage of haplogroup X in the Americas is not derived from a European subclade, but rather represent an independent subclade, labelled X2a.[24] The X2a subclade has not been found in Eurasia, and has most likely arisen within the early Paleo-Indian population, at roughly 13,000 years ago.[25] A basal variant of X2a was found in the Kennewick Man fossil (ca. 9,000 years ago).[26]
Subclades
Tree
This phylogenetic tree of haplogroup X subclades is based on the paper by Mannis van Oven and Manfred Kayser Updated comprehensive phylogenetic tree of global human mitochondrial DNA variation[3] and subsequent published research.
- X
- X1
- X1a
- X1a1
- X1b
- X1c
- X1a
- X2
- X2a
- X2a1
- X2a1a
- X2a1b
- X2a2
- X2a1
- X2b
- X2b1
- X2b2
- X2b3
- X2b4
- X2b5
- X2b6
- X2b7
- X2b8
- X2c
- X2c1
- X2c2
- X2d
- X2e
- X2e1
- X2e1a
- X2e1a1
- X2e1a1a
- X2e1a1
- X2e1a
- X2e2
- X2e2a
- X2e1
- X2f
- X2g
- X2h
- X2a
- X1
See also
- Human mitochondrial DNA haplogroups
- Indigenous American genetic studies
- Kennewick Man
- The Seven Daughters of Eve
|
Phylogenetic tree of human mitochondrial DNA (mtDNA) haplogroups | |||||||||||||||||||||||||||||||||||||||
| Mitochondrial Eve (L) | |||||||||||||||||||||||||||||||||||||||
| L0 | L1–6 | ||||||||||||||||||||||||||||||||||||||
| L1 | L2 | L3 | L4 | L5 | L6 | ||||||||||||||||||||||||||||||||||
| M | N | ||||||||||||||||||||||||||||||||||||||
| CZ | D | E | G | Q | O | A | S | R | I | W | X | Y | |||||||||||||||||||||||||||
| C | Z | B | F | R0 | pre-JT | P | U | ||||||||||||||||||||||||||||||||
| HV | JT | K | |||||||||||||||||||||||||||||||||||||
| H | V | J | T | ||||||||||||||||||||||||||||||||||||
References
- ↑ 1.0 1.1
X is estimated at 31.8+12.8
−12.1 kya (95% CI) in: "Supplementary Materials, Document S1" in: Soares, Pedro; Ermini, Luca; Thomson, Noel; Mormina, Maru; Rito, Teresa; Röhl, Arne; Salas, Antonio; Oppenheimer, Stephen et al. (12 June 2009). "Correcting for Purifying Selection: An Improved Human Mitochondrial Molecular Clock". American Journal of Human Genetics 84 (6): 740–759. doi:10.1016/j.ajhg.2009.05.001. PMID 19500773. - ↑ Havaš Auguštin, Dubravka; Šarac, Jelena; Reidla, Maere; Tamm, Erika; Grahovac, Blaženka; Kapović, Miljenko; Novokmet, Natalija; Rudan, Pavao et al. (August 2023). "Refining the Global Phylogeny of Mitochondrial N1a, X, and HV2 Haplogroups Based on Rare Mitogenomes from Croatian Isolates" (in en). Genes 14 (8): 1614. doi:10.3390/genes14081614. ISSN 2073-4425. PMID 37628665.
- ↑ 3.0 3.1 "Updated comprehensive phylogenetic tree of global human mitochondrial DNA variation". Human Mutation 30 (2): E386–394. Feb 2009. doi:10.1002/humu.20921. PMID 18853457.
- ↑ Bryan Sykes (2001). The Seven Daughters of Eve. London; New York: Bantam Press. ISBN 978-0-393-02018-2.
- ↑ Malhi, Ripan S.; Smith, David Glenn (September 2002). "Brief communication: Haplogroup X confirmed in prehistoric North America". American Journal of Physical Anthropology 119 (1): 84–86. doi:10.1002/ajpa.10106. PMID 12209576.
- ↑ Brown, Michael D.; Hosseini, Seyed H.; Torroni, Antonio; Bandelt, Hans-Jürgen; Allen, Jon C.; Schurr, Theodore G.; Scozzari, Rosaria; Cruciani, Fulvio et al. (1998-12-01). "mtDNA Haplogroup X: An Ancient Link between Europe/Western Asia and North America?". The American Journal of Human Genetics 63 (6): 1852–1861. doi:10.1086/302155. ISSN 0002-9297. PMID 9837837.
- ↑ Kujanová, Martina; Pereira, Luísa; Fernandes, Verónica; Pereira, Joana B.; Černý, Viktor (October 2009). "Near Eastern Neolithic genetic input in a small oasis of the Egyptian Western Desert". American Journal of Physical Anthropology 140 (2): 336–346. doi:10.1002/ajpa.21078. PMID 19425100.
- ↑ Perego, Ugo A.; Achilli, Alessandro; Angerhofer, Norman; Accetturo, Matteo; Pala, Maria; Olivieri, Anna; Kashani, Baharak Hooshiar; Ritchie, Kathleen H. et al. (January 2009). "Distinctive Paleo-Indian Migration Routes from Beringia Marked by Two Rare mtDNA Haplogroups". Current Biology 19 (1): 1–8. doi:10.1016/j.cub.2008.11.058. PMID 19135370.
- ↑ Lipson, Mark; Szécsényi-Nagy, Anna; Mallick, Swapan; Pósa, Annamária; Stégmár, Balázs; Keerl, Victoria; Rohland, Nadin; Stewardson, Kristin et al. (November 2017). "Parallel palaeogenomic transects reveal complex genetic history of early European farmers". Nature 551 (7680): 368–372. doi:10.1038/nature24476. PMID 29144465. Bibcode: 2017Natur.551..368L.
- ↑ Schuenemann, Verena J.; Peltzer, Alexander; Welte, Beatrix; van Pelt, W. Paul; Molak, Martyna; Wang, Chuan-Chao; Furtwängler, Anja; Urban, Christian et al. (August 2017). "Ancient Egyptian mummy genomes suggest an increase of Sub-Saharan African ancestry in post-Roman periods". Nature Communications 8 (1): 15694. doi:10.1038/ncomms15694. PMID 28556824. Bibcode: 2017NatCo...815694S.
- ↑ Fregel, Rosa; Méndez, Fernando L.; Bokbot, Youssef; Martín-Socas, Dimas; Camalich-Massieu, María D.; Santana, Jonathan; Morales, Jacob; Ávila-Arcos, María C. et al. (26 June 2018). "Ancient genomes from North Africa evidence prehistoric migrations to the Maghreb from both the Levant and Europe". Proceedings of the National Academy of Sciences of the United States of America 115 (26): 6774–6779. doi:10.1073/pnas.1800851115. PMID 29895688. Bibcode: 2018PNAS..115.6774F.
- ↑ 12.0 12.1 12.2 12.3 12.4 Reidla, Maere; Kivisild, Toomas; Metspalu, Ene; Kaldma, Katrin; Tambets, Kristiina; Tolk, Helle-Viivi; Parik, Jüri; Loogväli, Eva-Liis et al. (November 2003). "Origin and Diffusion of mtDNA Haplogroup X". The American Journal of Human Genetics 73 (5): 1178–1190. doi:10.1086/379380. PMID 14574647.
- ↑ Shlush, Liran I.; Behar, Doron M.; Yudkovsky, Guennady; Templeton, Alan; Hadid, Yarin; Basis, Fuad; Hammer, Michael; Itzkovitz, Shalev et al. (7 May 2008). "The Druze: A Population Genetic Refugium of the Near East". PLOS ONE 3 (5): e2105. doi:10.1371/journal.pone.0002105. PMID 18461126. Bibcode: 2008PLoSO...3.2105S.
- ↑ "Dolan DNA Learning Center – Native American haplogroups: European lineage, Douglas Wallace". Dnalc.org. http://www.dnalc.org/view/15188-Native-American-haplogroups-European-lineage-Douglas-Wallace.html.
- ↑ "The peopling of the Americas: Genetic ancestry influences health". Scientific American. http://www.physorg.com/news169474130.html.
- ↑ "Learn about Y-DNA Haplogroup Q" (Verbal tutorial possible). Wendy Tymchuk – Senior Technical Editor. Genebase Systems. 2008. http://www.genebase.com/tutorial/item.php?tuId=16.
- ↑ 17.0 17.1 Fagundes, Nelson J.R.; Kanitz, Ricardo; Eckert, Roberta; Valls, Ana C.S.; Bogo, Mauricio R.; Salzano, Francisco M.; Smith, David Glenn; Silva, Wilson A. et al. (March 2008). "Mitochondrial Population Genomics Supports a Single Pre-Clovis Origin with a Coastal Route for the Peopling of the Americas". The American Journal of Human Genetics 82 (3): 583–592. doi:10.1016/j.ajhg.2007.11.013. PMID 18313026.
- ↑ "An mtDNA view of the peopling of the world by Homo sapiens". Cambridge DNA Services. 2007. https://www.cambridgedna.com/genealogy-dna-ancient-migrations-slideshow.php?view=step7.
- ↑ Peñaloza-Espinosa, Rosenda I.; Arenas-Aranda, Diego; Cerda-Flores, Ricardo M.; Buentello-Malo, Leonor.; González-Valencia, Gerardo.; Torres, Javier; Álvarez, Berenice; Mendoza, Irma et al. (2007). "Characterization of mtDNA Haplogroups in 14 Mexican Indigenous Populations". Human Biology 79 (3): 313–320. doi:10.1353/hub.2007.0042. PMID 18078204.
- ↑ Easton, R. D.; Merriwether, D. A.; Crews, D. E.; Ferrell, R. E. (July 1996). "mtDNA variation in the Yanomami: evidence for additional New World founding lineages". American Journal of Human Genetics 59 (1): 213–225. PMID 8659527.
- ↑ "The presence of mitochondrial haplogroup x in Altaians from South Siberia". American Journal of Human Genetics 69 (1): 237–241. Jul 2001. doi:10.1086/321266. PMID 11410843.
- ↑ Bradley, Bruce; Stanford, Dennis (December 2004). "The North Atlantic ice-edge corridor: A possible Palaeolithic route to the New World". World Archaeology 36 (4): 459–478. doi:10.1080/0043824042000303656.
- ↑ Carey, Bjorn (19 February 2006)."First Americans may have been European.Life Science. Retrieved on 10 August 2007.
- ↑ "The similarities in ages and geographical distributions for C4c and the previously analyzed X2a lineage provide support to the scenario of a dual origin for Paleo-Indians. Taking into account that C4c is deeply rooted in the Asian portion of the mtDNA phylogeny and is indubitably of Asian origin, the finding that C4c and X2a are characterized by parallel genetic histories definitively dismisses the controversial hypothesis of an Atlantic glacial entry route into North America. "Mitochondrial haplogroup C4c: a rare lineage entering America through the ice-free corridor?". American Journal of Physical Anthropology 147 (1): 35–39. Jan 2012. doi:10.1002/ajpa.21614. PMID 22024980.
- ↑ X2a is dated 12.8+7.1
−6.7 kya in Soares et al. (2009). - ↑ "X2a has not been found anywhere in Eurasia, and phylogeography gives us no compelling reason to think it is more likely to come from Europe than from Siberia. Furthermore, analysis of the complete genome of Kennewick Man, who belongs to the most basal lineage of X2a yet identified, gives no indication of recent European ancestry and moves the location of the deepest branch of X2a to the West Coast, consistent with X2a belonging to the same ancestral population as the other founder mitochondrial haplogroups. Nor have any high-resolution studies of genome-wide data from Native American populations yielded any evidence of Pleistocene European ancestry or trans-Atlantic gene flow." Raff, Jennifer A.; Bolnick, Deborah A (2015). "Does Mitochondrial Haplogroup X Indicate Ancient Trans-Atlantic Migration to the Americas? A Critical Re-Evaluation". PaleoAmerica 1 (4): 297–304. doi:10.1179/2055556315Z.00000000040.
External links
- General
- Mannis van Oven's – mtDNA subtree N
- Haplogroup X
- Ian Logan's DNA Site
- Carolyn Benson's X mtDNA Project at Family Tree DNA
- Spread of Haplogroup X, from National Geographic
- CBC, The Solutrean Hypothesis, and Jennifer Raff, A podcast with Jennifer Raff discussions claims in 2018 linking Haplogroup X to the Solutrean hypothesis
Further reading
- "Heterogeneity of mitochondrial DNA haplotypes in Pre-Columbian Natives of the Amazon region". American Journal of Physical Anthropology 101 (1): 29–37. Sep 1996. doi:10.1002/(SICI)1096-8644(199609)101:1<29::AID-AJPA3>3.0.CO;2-8. PMID 8876812.
- Peter N. Jones (2004). American Indian mtDNA, Y Chromosome Genetic Data, and the Peopling of North America. Boulder: Bauu Press. ISBN 978-0-9721349-1-0. http://www.bauuinstitute.com/Publishing/DNAbook.html.
- "mtDNA variation in the Yanomami: evidence for additional New World founding lineages". American Journal of Human Genetics 59 (1): 213–225. Jul 1996. PMID 8659527.
- "mtDNA haplogroup X: An ancient link between Europe/Western Asia and North America?". American Journal of Human Genetics 63 (6): 1852–1861. Dec 1998. doi:10.1086/302155. PMID 9837837.
- "Distribution of mtDNA haplogroup X among Native North Americans". American Journal of Physical Anthropology 110 (3): 271–284. Nov 1999. doi:10.1002/(SICI)1096-8644(199911)110:3<271::AID-AJPA2>3.0.CO;2-C. PMID 10516561.
- "Mitochondrial DNA variation in the aboriginal populations of the Altai-Baikal region: implications for the genetic history of North Asia and America". Annals of the New York Academy of Sciences 1011 (1): 21–35. Apr 2004. doi:10.1196/annals.1293.003. PMID 15126280. Bibcode: 2004NYASA1011...21Z.
 |