Biology:Haplogroup M (mtDNA)
| Haplogroup M | |
|---|---|
 | |
| Possible time of origin | ca. 55,000-65,000 years ago[1] or 50,000-65,000 years ago[2] |
| Possible place of origin | South Asia,[3][4][5][6][7][8] Southwest Asia,[2][1] Southeast Asia,[9][10] or East Africa[11][12] |
| Ancestor | L3 |
| Descendants | M1, M2, M3, M4'45, M5, M6, M7, M8, M9, M10'42, M12'G, M13, M14, M15, M21, M27, M28, M29'Q, M31'32, M33, M34, M35, M36, M39, M40, M41, M44, M46, M47'50, M48, M49, M51, D |
| Defining mutations | 263, 489, 10400, 14783, 15043[13] |
Haplogroup M is a human mitochondrial DNA (mtDNA) haplogroup. An enormous haplogroup spanning all the continents, the macro-haplogroup M, like its sibling the macro-haplogroup N, is a descendant of the haplogroup L3.
All mtDNA haplogroups considered native outside of Africa are descendants of either haplogroup M or its sibling haplogroup N.[14] Haplogroup M is relatively young, having a younger most recent common ancestor date than some subclades of haplogroup N such as haplogroup R.[15]
Origins
There is a debate concerning the geographical origins of Haplogroup M and its sibling haplogroup N. Both lineages are thought to have been the main surviving lineages involved in the out of Africa migration (or migrations) because all indigenous lineages found outside Africa belong to haplogroup M or haplogroup N. Scientists are unsure whether the mutations that define haplogroups M and N occurred in Africa before the exit from Africa or in Asia after the exit from Africa. Determining the origins of haplogroup M is further complicated by an early back-migration (from Asia to Africa) of bearers of M1.[5]
Its date of origin in absolute terms is only known with great uncertainty, as reconstruction has yielded different (but overlapping) ranges for the age of M in South Asia and East Asia. The same authors give an estimate for t of L3 as 71.6+15.0
−14.5 kya,[16] later (2011) narrowed to the somewhat younger 65±5 kya.[1] Thus, haplogroup M would have emerged around 10,000 or at most 20,000 years after L3, around or somewhat after the recent out-of-Africa migration event.
Haplogroup M1
Much discussion concerning the origins of haplogroup M has been related to its subclade haplogroup M1, which is the only variant of macro-haplogroup M found in Africa.[14] Two possibilities were being considered as potential explanations for the presence of M1 in Africa:
- M was present in the ancient population which later gave rise to both M1 in Africa, and M more generally found in Eurasia.[17]
- The presence of M1 in Africa is the result of a back-migration from Asia which occurred sometime after the Out of Africa migration.[5]
Haplogroup M23
In 2009, two independent publications reported a rare, deep-rooted subclade of haplogroup M, referred to as M23, that is present in Madagascar .[18][19]
The contemporary populations of Madagascar were formed in the last 2,000 years by the admixture of Bantu and Indonesian (Austronesian) populations. M23 seems to be restricted to Madagascar, as it has not been detected anywhere else. M23 could have been brought to Madagascar from Asia where most deep rooted subclades of Haplogroup M are found.
Asian origin hypothesis
According to this theory, anatomically modern humans carrying ancestral haplogroup L3 lineages were involved in the Out of Africa migration from East Africa into Asia. Somewhere in Asia, the ancestral L3 lineages gave rise to haplogroups M and N. The ancestral L3 lineages were then lost by genetic drift as they are infrequent outside Africa. The hypothesis that Asia is the origin of macrohaplogroup M is supported by the following:
- The highest frequencies worldwide of macrohaplogroup M are observed in Asia, specifically in Bangladesh, China, India, Japan, Korea and Nepal where frequencies range from 60 to 80%. The total frequency of M subclades is even higher in some populations of Siberia or the Americas, but these small populations tend to exhibit strong genetic drift effects, and often their geographical neighbors exhibit very different frequencies.[3][20][21]
- Deep time depth >50,000 years of western, central, southern and eastern Indian haplogroups M2, M38, M54, M58, M33, M6, M61, M62 and the distribution of macrohaplogroup M, do not rule out the possibility of macrohaplogroup M arising in Indian population.[22]
- With the exception of the African specific M1, India has several M lineages that emerged directly from the root of haplogroup M.[3][21]
- Only two subclades of haplogroup M, M1 and M23, are found in Africa, whereas numerous subclades are found outside Africa[3][5] (with some discussion possible only about sub-clade M1, concerning which see below).
- Specifically concerning M1
- Haplogroup M1 has a restricted geographic distribution in Africa, being found mainly in North Africans and East Africa at low or moderate frequencies. If M had originated in Africa around, or before, the Out of Africa migration, it would be expected to have a more widespread distribution [21]
- According to Gonzalez et al. 2007, M1 appears to have expanded relatively recently. In this study M1 had a younger coalescence age than the Asian-exclusive M lineages.[5]
- The geographic distribution of M1 in Africa is predominantly North African/supra-equatorial[5] and is largely confined to Afro-Asiatic speakers,[23] which is inconsistent with the Sub-Saharan distribution of sub-clades of haplogroups L3 and L2 that have similar time depths.[14]
- One of the basal lineages of M1 lineages has been found in Northwest Africa and in the Near East but is absent in East Africa.[5]
- M1 is not restricted to Africa. It is relatively common in the Mediterranean, peaking in Iberia. M1 also enjoys a well-established presence in the Middle East, from the South of the Arabian Peninsula to Anatolia and from the Levant to Iran. In addition, M1 haplotypes have occasionally been observed in the Caucasus and the Trans Caucasus, and without any accompanying L lineages.[5][14] M1 has also been detected in Central Asia, seemingly reaching as far as Tibet.[5]
- The fact that the M1 sub-clade of macrohaplogroup M has a coalescence age which overlaps with that of haplogroup U6 (a Eurasian haplogroup whose presence in Africa is due to a back-migration from West Asia) and the distribution of U6 in Africa is also restricted to the same North African and Horn African populations as M1 supports the scenario that M1 and U6 were part of the same population expansion from Asia to Africa.[23]
- The timing of the proposed migration of M1 and U6-carrying peoples from West Asia to Africa (between 40,000 to 45,000 ybp) is also supported by the fact that it coincides with changes in climatic conditions that reduced the desert areas of North Africa, thereby rendering the region more accessible to entry from the Levant. This climatic change also temporally overlaps with the peopling of Europe by populations bearing haplogroup U5, the European sister clade of haplogroup U6.[23]
African origin hypothesis
According to this theory, haplogroups M and N arose from L3 in an East African population ancestral to eurasians that had been isolated from other African populations before the OOA event. Members of this population were involved in the out Africa migration and may have only carried M and N lineages. With the possible exception of haplogroup M1, all other M and N clades in Africa were lost due to admixture with other African populations and genetic drift.[8][17]
The African origin of Haplogroup M is supported by the following arguments and evidence.
- L3, the parent clade of haplogroup M, is found throughout Africa, but is rare outside Africa.[17] According to Toomas Kivisild (2003), "the lack of L3 lineages other than M and N in India and among non-African mitochondria in general suggests that the earliest migration(s) of modern humans already carried these two mtDNA ancestors, via a departure route over the Horn of Africa."[8]
- Specifically concerning at least M1a:
- This study provides evidence that M1, or its ancestor, had an Asiatic origin. The earliest M1 expansion into Africa occurred in northwestern instead of northeastern areas; this early spread reached the Iberian Peninsula even affecting the Basques. The majority of the M1a lineages found outside and inside Africa had a more recent eastern Africa origin. Both western and eastern M1 lineages participated in the Neolithic colonization of the Sahara. The striking parallelism between subclade ages and geographic distribution of M1 and its North African U6 counterpart strongly reinforces this scenario. Finally, a relevant fraction of M1a lineages present today in the European Continent and nearby islands possibly had a Jewish instead of the commonly proposed Arab/Berber maternal ascendance.[5]
Dispersal

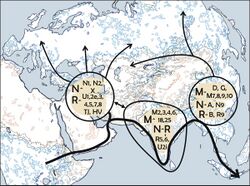
A number of studies have proposed that the ancestors of modern haplogroup M dispersed from Africa through the southern route across the Horn of Africa along the coastal regions of Asia onwards to New Guinea and Australia. These studies suggested that the migrations of haplogroups M and N occurred separately with haplogroup N heading northwards from East Africa to the Levant. However, the results of numerous recent studies indicate that there was only one migration out of Africa and that haplogroups M and N were part of the same migration. This is based on the analysis of a number of relict populations along the proposed beachcombing route from Africa to Australia, all of which possessed both haplogroups N and M.[4][24]
A 2008 study by Abu-Amero et al., suggests that the Arabian Peninsula may have been the main route out of Africa. However, as the region lacks of autochthonous clades of haplogroups M and N the authors suggest that the area has been a more recent receptor of human migrations than an ancient demographic expansion center along the southern coastal route as proposed under the single migration Out-of-Africa scenario of the African origin hypothesis.[6]
Distribution
M is the most common mtDNA haplogroup in Asia,[25] super-haplogroup M is distributed all over Asia, where it represents 60% of all maternal lineages.[26]
All Andamanese belong to Haplogroup M.[27] It peaks in the Malaysian aboriginal Negrito tribes at almost 100% but with mtDNA M21a representing Semang; 84% in Mendriq people, Batek people 48%, (almost all belong to the specific Malaysian Negrito haplogroup M21a, this subclade also found in the Orang Asli 21%, Thais 7.8% and Malay 4.6%)[28][29][30] It also peaks very high in Japan and Tibet, where it represents on average about 70% of the maternal lineages (160/216 = 74% Tibet,[31] 205/282 = 73% Tōkai,[32] 231/326 = 71% Okinawa,[32] 148/211 = 70% Japanese,[20] 50/72 = 69% Tibet,[31] 150/217 = 69% Hokkaidō,[33] 24/35 = 69% Zhongdian Tibetan, 175/256 = 68% northern Kyūshū,[32] 38/56 = 68% Qinghai Tibetan, 16/24 = 67% Diqing Tibetan, 66/100 = 66% Miyazaki, 33/51 = 65% Ainu, 214/336 = 64% Tōhoku,[32] 75/118 = 64% Tokyo (JPT)[34]) and is ubiquitous in India[3][14][35] and South Korea ,[32][36][37][38][39] where it has approximately 60% frequency. Among Chinese people both inside and outside of China, haplogroup M accounts for approximately 50% of all mtDNA on average, but the frequency varies from approximately 40% in Hans from Hunan and Fujian in southern China to approximately 60% in Shenyang, Liaoning in northeastern China.[31][32][34][37]
Haplogroup M accounts for approximately 42% of all mtDNA in Filipinos, among whom it is represented mainly by M7c3c and E.[40] In Vietnam, haplogroup M has been found in 37% (52/139) to 48% (20/42) of samples of Vietnamese and in 32% (54/168) of a sample of Chams from Bình Thuận Province.[37][41] Haplogroup M accounts for 43% (92/214) of all mtDNA in a sample of Laotians, with its subclade M7 (M7b, M7c, and M7e) alone accounting for a full third of all haplogroup M, or 14.5% (31/214) of the total sample.[42]
In Oceania, A 2008 study found Haplogroup M in 42% (60/144) of a pool of samples from nine language groups in the Admiralty Islands of Papua New Guinea.,[43] M has been found in 35% (17/48) of a sample of Papua New Guinea highlanders from the Bundi area and in 28% (9/32) of a sample of Aboriginal Australians from Kalumburu in northwestern Australia.[44] In a study published in 2015, Haplogroup M was found in 21% (18/86) of a sample of Fijians, but it was not observed in a sample of 21 Rotumans.[45]
Haplogroup M is also relatively common in Northeast Africa, occurring especially among Somalis, Libyans and Oromos at frequencies over 20%.[46][47] Toward the northwest, the lineage is found at comparable frequencies among the Tuareg in Mali and Burkina Faso; particularly the M1a2 subclade (18.42%).[48]
Among the descendant lineages of haplogroup M are C, D, E, G, Q, and Z. Z and G are found in North Eurasian populations, C and D exists among North Eurasian and Native American populations, E is observed in Southeast Asian populations, and Q is common among Melanesian populations. The lineages M2, M3, M4, M5, M6, M18 and M25 are exclusive to South Asia, with M2 reported to be the oldest lineage on the Indian sub-continent with an age estimation of 60,000—75,000 years, and with M5 reported to be the most prevalent in historically Turco-Persian enclaves.[3][49][50]
In 2013, four ancient specimens dated to around 2,500 BC-500 AD, which were excavated from the Tell Ashara (Terqa) and Tell Masaikh (Kar-Assurnasirpal) archaeological sites in the Euphrates Valley, were found to belong to mtDNA haplotypes associated with the M4b1, M49 and/or M61 haplogroups. Since these clades are not found among the current inhabitants of the area, they are believed to have been brought at a more remote period from east of Mesopotamia; possibly by either merchants or the founders of the ancient Terqa population.[51]
In 2016, three Late Pleistocene European hunter-gatherers were also found to carry M lineages. Two of the specimens were from the Goyet archaeological site in Belgium and were dated to 34,000 and 35,000 years ago, respectively. The other ancient individual hailed from the La Rochette site in France, and was dated to 28,000 years ago.[52]
Ancient DNA analysis of Iberomaurusian skeletal remains at the Taforalt site in Morocco, which have been dated to between 15,100 and 13,900 ybp, observed the M1b subclade in one of the fossils (1/7; ~14%).[53] Ancient individuals belonging to the Late Iron Age settlement of Çemialo Sırtı in Batman, southeast Turkey were found to carry haplogroup M; specifically the M1a1 subclade (1/12; ~8.3%). Haplogroup M was also detected in ancient specimens from Southeast Anatolia (0.4%).[54] Additionally, M1 has been observed among ancient Egyptian mummies excavated at the Abusir el-Meleq archaeological site in Middle Egypt, which date from the Pre-Ptolemaic/late New Kingdom and Roman periods.[55] Fossils at the Early Neolithic site of Ifri n'Amr or Moussa in Morocco, which have been dated to around 5,000 BCE, have also been found to carry the M1b subclade. These ancient individuals bore an autochthonous Maghrebi genomic component that peaks among modern Berbers, indicating that they were ancestral to populations in the area.[56] The ancient Egyptian aristocrats Nakht-Ankh and Khnum-Nakht were also found to belong to the M1a1 subclade. The half-brothers lived during the 12th Dynasty, with their tomb located at the Deir Rifeh cemetery in Middle Egypt.[57]
Subgroups distribution
Source:[58]
- Haplogroup M1 [2] – found in the Nile Valley, Horn of Africa, North Africa, Sahara, Mediterranean, and Middle East[3][5]
- M20 – in China,[59] Borneo (Bidayuh[60]), Thailand, Laos, Indonesia, Vietnam (Jarai, Lahu), Saudi Arabia
- M20a - Myanmar, Thailand (Shan from Mae Hong Son Province, Tai Yuan), China, Blang, Saudi Arabia,
- M51 – in Cambodia[63] and Indonesia
- Haplogroup M2 [3] – found in South Asia, with highest concentrations in SE India and Bangladesh;[14] oldest haplogroup M lineage on the Indian sub-continent.[3] Also found with low frequency in southwestern China.[31]
- M2a'b
- M2a – India (Madhya Pradesh), Munda; most common in Bangladesh
- M2a1 – Malpaharia
- M2a1a – Uyghur, Sindhi, Hill Kolam, Thailand
- M2a1a1 – Katkari
- M2a1a1a – Nihal
- M2a1a1a1 – Katkari
- M2a1a1b – Nihal
- M2a1a1b1 – Nihal
- M2a1a1a – Nihal
- M2a1a2 – Madia
- M2a1a2a – Madia
- M2a1a2a1 – Kamar
- M2a1a2a1a – Kamar
- M2a1a2a1 – Kamar
- M2a1a2a – Madia
- M2a1a3 – Mathakur
- M2a1a3a – Kathakur, Mathakur
- M2a1a3a1 – Kathakur
- M2a1a3b – Kathakur
- M2a1a3a – Kathakur, Mathakur
- M2a1a1 – Katkari
- M2a1b – Dungri Bhil
- M2a1c – Andh
- M2a1a – Uyghur, Sindhi, Hill Kolam, Thailand
- M2a1 – Malpaharia
- M2b – Paudi Bhuiya; most common in SE India
- M2b1 – Korku
- M2b1a – Korku, Munda
- M2b1b – Malpaharia
- M2b2 – Hill Kolam, Jenu Kuruba
- M2b3 – Betta Kurumba
- M2b3a – Betta Kurumba
- M2b4 – Korku
- M2b1 – Korku
- M2c – Myanmar, Thailand/Laos
- M2a – India (Madhya Pradesh), Munda; most common in Bangladesh
- M2a'b
- Haplogroup M3 – Uyghur, Myanmar, New Delhi (Hindu), Paniya [4] – found mainly in South Asia, with highest concentrations in west and NW India[14]
- M3a
- M3a1 – Dongri Bhil, Kathodi, Katkari, Jammu and Kashmir, Pathan, Iran, Thailand/Laos
- M3a2 – Bangladesh, Jammu and Kashmir, Burusho, Qatar, Yemen
- M3a2a – Jenu Kuruba
- M3b – Kamar of Chhattisgarh
- M3c – Madia, Myanmar
- M3c1
- M3c1a – Jammu and Kashmir, Nepal (Terai Hindu, Tharu), Andhra Pradesh (tribal)
- M3c1b – Hill Kolam
- M3c1b1 – Saudi Arabia
- M3c1b1a – Jenu Kuruba
- M3c1b1b – Jenu Kuruba
- M3c1b1 – Saudi Arabia
- M3c2 – Pakistan (Brahui), Jammu and Kashmir, Andh, Thailand
- M3c1
- M3d – Nepal (Kathmandu), India, Italy (Salerno)
- M3d1 – New Delhi (Hindu)
- M3d1a – Nepal (Kathmandu), Cambodia (Lao), United Kingdom
- M3d1a1 – Tibet (Sherpa)
- M3d1a – Nepal (Kathmandu), Cambodia (Lao), United Kingdom
- M3d1 – New Delhi (Hindu)
- M3a
- M4'30
- Haplogroup M4 [5] – found mainly in South Asia but some sequences in Eastern Saudi Arabia
- Haplogroup M4a – found in Gujarat, India[21]
- Haplogroup M4b – found among ancient specimens in the Euphrates valley
- Haplogroup M65
- Haplogroup M30 – mainly in India; also found in Nepal, Pakistan, Central Asia (Kyrgyz, Wakhi, and Sarikoli in Taxkorgan, Xinjiang, China and Tajiks in Dushanbe, Tajikistan[50]), the Middle East, and North Africa.
- Haplogroup M18'38
- Haplogroup M18 – found among Tharus in southern Nepal and tribal people in Andhra Pradesh[65] Haplogroup M18a was also found in Mesolithic Sri Lanka.[66]
- Haplogroup M38 – found with high frequency among Tharus from Morang District of southeastern Nepal and as singletons among Tharus from Chitwan District of south-central Nepal and Hindus from New Delhi[65]
- Haplogroup M18'38
- Haplogroup M37
- Haplogroup M37a – found in Gujarat, India[21]
- Haplogroup M4 [5] – found mainly in South Asia but some sequences in Eastern Saudi Arabia
- Haplogroup M5 [6] – found in South Asia
- Haplogroup M5a – found in India (Jammu and Kashmir, Madhya Pradesh, Kathakur,[67] Gadaba[21]), Thailand (Mon in Ratchaburi Province and Lopburi Province[58]), Israel, Kyrgyz in Taxkorgan[50]
- M5a1
- M5a1a – India (incl. Jammu and Kashmir)
- M5a1b – India (Jammu and Kashmir, Dongri Bhil, Nihal, Andh), Pakistan (Burusho), Russia,[68] Spain (Romani), USA (Georgia), USA (California)
- M5a2 – India
- M5a2a – Pakistan (Balochi), India (Nihal), Thailand (Tai Yuan in Uttaradit Province[58])
- M5a2a1 – India (Hindus in New Delhi), Pakistan (Sindhi)
- M5a2a1a – Saudi Arabia, Iran (Persian),[49] Kazakh, Pakistan (Balochi), India (Dongri Bhil, Korku, Lachungpa), Myanmar
- M5a2a2 – India (Kamar of Chhattisgarh), Yemen
- M5a2a3 – India (Pauri Bhuiya, Munda)
- M5a2a4 – Iran (Persians),[49] Pakistan (Brahui, Makrani, Balochi)
- M5a2a1 – India (Hindus in New Delhi), Pakistan (Sindhi)
- M5a2a – Pakistan (Balochi), India (Nihal), Thailand (Tai Yuan in Uttaradit Province[58])
- M5a3
- M5a4 – India (Kathodi,[67] Korku[67])
- M5a5 – India (Dongri Bhil,[67] Andh[67]), Yemen
- M5a1
- Haplogroup M5b – found in India and Thailand (Khon Mueang in Chiang Mai Province[58])
- Haplogroup M5b2b1a – found in Tibet, Ladakh, Nepal
- Haplogroup M5c – found in India, Thailand (Mon in Lopburi Province and Nakhon Ratchasima Province[58]), Tibet, Nepal
- Haplogroup M5a – found in India (Jammu and Kashmir, Madhya Pradesh, Kathakur,[67] Gadaba[21]), Thailand (Mon in Ratchaburi Province and Lopburi Province[58]), Israel, Kyrgyz in Taxkorgan[50]
- Haplogroup M6 [7] – found mainly in South Asia, with highest concentrations in mid-eastern India and Kashmir[14]
- Haplogroup M6b – found in Kerala, India[21]
- Haplogroup M61 – found among ancient specimens in the Euphrates valley
- Haplogroup M7 [8] – found in East Asia and Southeast Asia, especially in Japan, southern China, Vietnam,[70] Laos,[42] and Thailand;[58] also found with low frequency in Central Asia and Siberia
- Haplogroup M7a
- Haplogroup M7a* – Japan
- Haplogroup M7a1
- Haplogroup M7a1* – Japan, Jiangsu, Shandong
- Haplogroup M7a1a
- Haplogroup M7a1a* – Japan, Korea, Beijing, Hebei, Henan, Sichuan, Shanghai, Shandong
- Haplogroup M7a1a1
- Haplogroup M7a1a1* – Japan, Korea, Jiangsu, Shandong, Liaoning, Henan
- Haplogroup M7a1a1a – Japan
- Haplogroup M7a1a2
- Haplogroup M7a1a2* – Japan, Jiangsu
- Haplogroup M7a1a2a – Japan
- Haplogroup M7a1a3 – Japan
- Haplogroup M7a1a4
- Haplogroup M7a1a4* – Japan, Zhejiang
- Haplogroup M7a1a4a – Japan
- Haplogroup M7a1a5
- Haplogroup M7a1a5* – Japan
- Haplogroup M7a1a5a – Japan, Korea, Tianjin
- Haplogroup M7a1a6
- Haplogroup M7a1a6* – Japan, Philippines, Jiangsu, Shanxi, Shandong
- Haplogroup M7a1a6a – Japan
- Haplogroup M7a1a7
- Haplogroup M7a1a7* – Japan, Korea
- Haplogroup M7a1a7a – Uyghur
- Haplogroup M7a1a8 – Japan, Jiangsu
- Haplogroup M7a1a9 – Japan, Korea, Tianjin
- Haplogroup M7a1a10 – Japan
- Haplogroup M7a1b
- Haplogroup M7a1b1
- Haplogroup M7a1b1* – Japan, China (Minnan Han)
- Haplogroup M7a1b1a – Japan
- Haplogroup M7a1b2 – Japan
- Haplogroup M7a1b1
- Haplogroup M7a2
- Haplogroup M7b'c
- Haplogroup M7b
- Haplogroup M7b1a
- Haplogroup M7b1a1 – Thailand, Laos, Vietnam, Cambodia, Myanmar, Indonesia, China, Karakalpak, Kyrgyz, Mongush, Khamnigan
- Haplogroup M7b1a1a – Thailand, Uyghur, Korea
- Haplogroup M7b1a1a1 – Japan, Korea, China, Tajikistan, Thailand, Laos
- Haplogroup M7b1a1a1a – Japan
- Haplogroup M7b1a1a1b – Japan, Korea, China, Russia, Kyrgyzstan
- Haplogroup M7b1a1a1c – Japan
- Haplogroup M7b1a1a1d – Japan
- Haplogroup M7b1a1a2 – Thailand, Vietnam, Malaysia, China
- Haplogroup M7b1a1a3 – Thailand, Laos, Vietnam, China
- Haplogroup M7b1a1a1 – Japan, Korea, China, Tajikistan, Thailand, Laos
- Haplogroup M7b1a1b – Thailand, Laos, Vietnam, China (Hunan, Uyghur, Kyrgyz from Artux[50]), England
- Haplogroup M7b1a1c – Han Chinese, Uyghurs, Kyrgyz
- Haplogroup M7b1a1c1 – Chinese, Bama Yao Autonomous County
- Haplogroup M7b1a1d – Thailand, Laos, Tatar (Buinsk)
- Haplogroup M7b1a1e – Thailand
- Haplogroup M7b1a1e1 – Thailand, Vietnam, China
- Haplogroup M7b1a1e2 – China
- Haplogroup M7b1a1f – Thailand, Malaysia, Indonesia, Vietnam, China
- Haplogroup M7b1a1g – Thailand
- Haplogroup M7b1a1h – Thailand, Chinese (Han from Lanzhou,[74] etc.), Vietnam, Korea, Japan
- Haplogroup M7b1a1i – Taiwan (Amis), Philippines, Malaysia
- Haplogroup M7b1a1a – Thailand, Uyghur, Korea
- Haplogroup M7b1a2
- Haplogroup M7b1a2a – China (Uyghurs, Kyrgyzes in Taxkorgan, Han, Mongol in Inner Mongolia)
- Haplogroup M7b1a2a1 – Taiwan Aboriginal peoples, Philippines, Indonesia, Malaysia
- Haplogroup M7b1a2a1a – Atayal, Saisiyat
- Haplogroup M7b1a2a1b – Atayal
- Haplogroup M7b1a2a1b1 – Atayal, Saisiyat
- Haplogroup M7b1a2a1 – Taiwan Aboriginal peoples, Philippines, Indonesia, Malaysia
- Haplogroup M7b1a2a – China (Uyghurs, Kyrgyzes in Taxkorgan, Han, Mongol in Inner Mongolia)
- Haplogroup M7b1a1 – Thailand, Laos, Vietnam, Cambodia, Myanmar, Indonesia, China, Karakalpak, Kyrgyz, Mongush, Khamnigan
- Haplogroup M7b1b – Khamnigan, China, Kyrgyz
- Haplogroup M7b1a
- Haplogroup M7c
- Haplogroup M7c1 – China, Vietnam, Malaysia, Mongolia, Sarikoli, Kazakhstan
- Haplogroup M7c1a – China, Korea, Japan, Vietnam, Thailand, Indonesia
- Haplogroup M7c1a1
- Haplogroup M7c1a1a – China, Mongolia
- Haplogroup M7c1a1b – Azeri
- Haplogroup M7c1a1b1 – Even (Sakkyryyr), Yakut (Vilyuy basin)
- Haplogroup M7c1a2 – China
- Haplogroup M7c1a2a – She, Uyghur
- Haplogroup M7c1a2a1 – Japan, Korea, Uyghur
- Haplogroup M7c1a2a – She, Uyghur
- Haplogroup M7c1a3 – China, Japan, Vietnam
- Haplogroup M7c1a3a – Korea
- Haplogroup M7c1a4
- Haplogroup M7c1a4a – China
- Haplogroup M7c1a4b – China
- Haplogroup M7c1a5 – Japan, Korea
- Haplogroup M7c1a1
- Haplogroup M7c1b – Chinese
- Haplogroup M7c1b1 – Buryats
- Haplogroup M7c1b2
- Haplogroup M7c1b2a – Khamnigan, Korea
- Haplogroup M7c1b2b – China, Thailand, Laos, Malaysia
- Haplogroup M7c1c – China, Thailand/Laos
- Haplogroup M7c1c1 – China
- Haplogroup M7c1c1a – China
- Haplogroup M7c1c1a1 – Philippines
- Haplogroup M7c1c1a – China
- Haplogroup M7c1c2 – Thailand, China (Han)
- Haplogroup M7c1c3 – Taiwan, Thailand, Philippines, Indonesia, Brunei, Malaysia, Kiribati, Nauru, Saudi Arabia, Madagascar
- Haplogroup M7c1c1 – China
- Haplogroup M7c1a – China, Korea, Japan, Vietnam, Thailand, Indonesia
- Haplogroup M7c2 – Taiwan, Hainan, Thailand/Laos
- Haplogroup M7c2a – Thailand/Laos, China (incl. Hainan)
- Haplogroup M7c2b – Thailand, Taiwan (Han), Czech
- Haplogroup M7c3 – China (incl. Amis)
- Haplogroup M7c1 – China, Vietnam, Malaysia, Mongolia, Sarikoli, Kazakhstan
- Haplogroup M7b
- Haplogroup M7a
- Haplogroup M8 - China,[61] Northern Thailand (Lisu[62][61]), India
- Haplogroup M8a: [9] – found in East Asia, Central Asia, and Siberia
- Haplogroup M8a1
- Haplogroup M8a2'3
- Haplogroup M8a2'3* – Japan
- Haplogroup M8a2 – found in Koryaks, Itelmens, Chukchis, Tuvans, Khakassians, Altayans, Mongolians, China (including Uyghurs), Koreans, Japan, Thailand/Laos[38][75]
- Haplogroup M8a2* – China (Hakka)
- Haplogroup M8a2-T152C!!!
- Haplogroup M8a2-T152C!!!* – China, Japan (Chiba)[61]
- Haplogroup M8a2a
- Haplogroup M8a2a1 – found in Thailand, China, Japan
- Haplogroup M8a2a1a
- Haplogroup M8a2a1a* – Northeast Thailand (Saek)
- Haplogroup M8a2a1a1
- Haplogroup M8a2a1a1* – China (Han from Wuhan)
- Haplogroup M8a2a1a1a – Central Thailand (Tai Yuan), Northern Thailand (Palaung)
- Haplogroup M8a2a1b
- Haplogroup M8a2a1b* – Chiang Mai Province (Khon Mueang)
- Haplogroup M8a2a1b1 – Lamphun Province (Khon Mueang)
- Haplogroup M8a2a1c – China, Japan (Aichi)
- Haplogroup M8a2a1a
- Haplogroup M8a2a2 – China
- Haplogroup M8a2a1 – found in Thailand, China, Japan
- Haplogroup M8a2-A12530G/G14364A/T16297C – Uyghur
- Haplogroup M8a2b – China (Maonan from Pingtang County[76])
- Haplogroup M8a2b* - Japan,[61] China (Shandong,[61] Henan[61])
- Haplogroup M8a2b1 (C9301T) - China (Anhui Medical University Hospital,[61] etc.)
- Haplogroup M8a2b2 (G12192A) - Russia (Ulchi,[61] Nanai from Bogorodskoye, Ulchsky District[61])
- Haplogroup M8a2b3 (T16311C!) - China,[61] Thailand (Bangkok[61])
- Haplogroup M8a2b4 (T9813d) - China (Zhejiang, etc.[61])
- Haplogroup M8a2c – found in Japan and China
- Haplogroup M8a2d – found in China (Shantou, Qingdao)
- Haplogroup M8a2e – found in Taiwan (Ami, etc.) and in a Han Chinese living in the Denver, Colorado metropolitan area
- Haplogroup M8a2f – China
- Haplogroup M8a3
- Haplogroup M8a3* – China (Guangdong, etc.), Kyrgyz (Artux[50]), Russia (Verkhnyaya Gutara, Nerkha, and Kushun villages of Irkutsk Oblast)
- Haplogroup M8a3a
- Haplogroup M8a3a* – China, Russia (Ket from Turukhansk)
- Haplogroup M8a3a1
- Haplogroup M8a3a1* – China
- Haplogroup M8a3a1a – China
- Haplogroup M8a3a2 – China, Indonesia (Jawa Timur)
- Haplogroup CZ - Northern Thailand (Hmong[62])
- Haplogroup C [10] – found especially in Siberia
- Haplogroup Z [14] – found in Northeast Europe, Siberia, Central Asia, and East Asia, including among Swedes, Sami, Finns, Russians, Ukrainians, Nogais, Abazins, Cherkessians, North Ossetians, Turks, Udmurts, Komi, Kets, Kalmyks, Hazara, Pashtuns, Tajiks, Turkmens, Uzbeks, Kazakhs, Kyrgyz, Uyghurs, Evens, Evenks, Dolgans, Yakuts, Yukaghirs, Khakas, Altaians, Altai Kizhi, Buryats, Nganasans, Koryaks, Itelmens, Ulchi, Japanese, Koreans, Chinese, and Tibeto-Burman peoples
- Haplogroup M8a: [9] – found in East Asia, Central Asia, and Siberia
- Haplogroup M9 [15] – found in East Asia and Central Asia, especially in Tibet. In the Nepalese populations, it is prevalent mainly in Sherpa (27.4%), Tharu-CI (19.6%), Tamang (15.5%), Magar (13.5%), and Tharu-CII.[77] Haplogroup M9* has additionally been found in ancient remains from the Red Deer Cave people in present-day Yunnan.[78]
- Haplogroup M9a'b
- Haplogroup M9a – Han (Guangdong, Guangxi, Yunnan, Sichuan, Hunan, Taiwan, Anhui, Shaanxi, Shandong, Hebei), Korean (South Korea), Tujia (Hunan), Kinh (Hue), Mongol (Hohhot), Japanese, Lhoba[61] [TMRCA 23,000 (95% CI 18,100 <-> 28,800) ybp[61]]
- Haplogroup M9a1 – Han (Hunan) [TMRCA 19,500 (95% CI 13,800 <-> 26,700) ybp[61]]
- Haplogroup M9a1a – Han (Hebei, Henan, Shaanxi, Anhui, Zhejiang, Hunan, Yunnan, Guangdong, Hong Kong, Taiwan), Manchu (Jilin), Korean (South Korea), Hui (Qinghai), Kazakh (Ili), Kyrgyz (Kyrgyzstan), Nepal [TMRCA 16,500 (95% CI 12,800 <-> 20,900) ybp[61]]
- Haplogroup M9a1a1 – Han (Henan, Shaanxi, Guangdong, Guangxi, Sichuan, Yunnan), Taiwan, Thailand/Laos, Hui (Yuxi), Tibetan (Nyingchi), Uyghur, Japanese (Hokkaido) [TMRCA 13,900 (95% CI 10,800 <-> 17,600) ybp[61]]
- Haplogroup M9a1a1a – Japanese, Korean (Seoul), Chinese (incl. a Henan Han), Khamnigan (Buryat Republic), Udege, Nivkh, Tibetan (Qinghai)
- Haplogroup M9a1a1b – Japanese, Korean (South Korea), Mongol (Inner Mongolia), Han (Hunan)
- Haplogroup M9a1a1c – Han (Gansu, Shaanxi, Henan, Liaoning, Zhejiang, Jiangxi, Hunan, Guangdong, Sichuan, Yunnan), Ainu, Japanese, Korean, Mongol (Hohhot), Uyghur (Ürümqi), Altaian, Tuvinian, Hui (Xinjiang, Kyrgyzstan), Tujia (Hunan), Bai (Yunnan), Yi (Yunnan)
- Haplogroup M9a1a1c1 – Han (Henan)
- Haplogroup M9a1a1c1a – Han (Henan, Anhui, Shandong, Liaoning, Sichuan, Yunnan, Xinjiang, Lanzhou[74]), Korea, Japanese, Mongol (New Barga Left Banner), Tibetan (Liangshan), Hui (Ili)
- Haplogroup M9a1a1c1b – Tibetan (Gansu, Qinghai, Sichuan, Yunnan, Chamdo, Lhasa, Nagqu, Ngari, Nyingchi, Shannan, Shigatse), Monpa (Nyingchi), Dirang Monpa (Arunachal Pradesh), Lachungpa (Sikkim), Tu (Huzhu Tu Autonomous County), Dongxiang (Gansu), Buryat (Inner Mongolia, Buryat Republic), Han (Qinghai), Hui (Qinghai), Nepalese
- Haplogroup M9a1a1c1 – Han (Henan)
- Haplogroup M9a1a1d – Salar (Qinghai), Han (Yanting), Bai (Dali)
- Haplogroup M9a1a2 – Tharu (Chitwan District, Uttar Pradesh), Tibetan (Nagqu, Yunnan, Qinghai, Shigatse), Lhoba (Nyingchi), Dhimal (West Bengal), Chin (Myanmar), Adi (Assam), Tu (Qinghai), Uyghur (Ürümqi), Mongol (Ili), Han (Hunan, Shanxi, Sichuan, Yunnan, Shandong, Ili), Yi (Yunnan), Bai (Dali), Nepalese [TMRCA 6,153.9 ± 5,443.2 ybp; CI=95%[79]]
- Haplogroup M9a1a1 – Han (Henan, Shaanxi, Guangdong, Guangxi, Sichuan, Yunnan), Taiwan, Thailand/Laos, Hui (Yuxi), Tibetan (Nyingchi), Uyghur, Japanese (Hokkaido) [TMRCA 13,900 (95% CI 10,800 <-> 17,600) ybp[61]]
- Haplogroup M9a1b – Tibetan (Nyingchi, Nagqu, Lhasa, Chamdo, Ngari, Shannan, Shigatse, Sichuan, Yunnan, Qinghai, Gansu), Monba (Nyingchi), Lhoba (Shannan), Uzbekistan (Fergana), Dongxiang (Linxia), Naga (Sagaing), Burman (Bago), Chin (Chin State), Han (Hunan, Sichuan, Yunnan, Liaoning), Yi (Shuangbai) [TMRCA 9,416.6 ± 3,984.0 ybp; CI=95%[79]]
- Haplogroup M9a1b1 – Tibetan, Lhoba, Arunachal Pradesh (Sonowal Kachari, Wanchoo, Gallong), Assam (Adi), Sikkim (Lepcha, Lachung), Qinghai (Salar, Tu), Mongol (Mongolia, Inner Mongolia), Guangxi (Gelao, Palyu), Thailand, Bengal, Pakistan (Karachi), Meghalaya (Khasi, Garo), Bodo (West Bengal), Rabha (West Bengal), Rajbanshi (West Bengal), Indonesia, Han (Shaanxi, Henan, Gansu, Sichuan, Yunnan), Hui (Gansu, Qinghai), Burman (Ayeyarwady, Magway, Sagaing), Rakhine (Rakhine, Magway), Chin (Chin State), Naga (Sagaing), Mech (Jhapa district, Nepal), Nepalese, Mosuo (Yunnan), Yi (Yunnan), She (Guizhou), Hani (Yunnan), Pumi (Yunnan), Bai (Dali), Va (Yunnan) [TMRCA 6,557.4 ± 2,102.4 ybp; CI=95%[79]]
- Haplogroup M9a1b2 – Tibetan (Diqing), Han (Dujiangyan), Kazakh (Altai Republic), Kalmyk [TMRCA 3,225.9 ± 3,494.4 ybp; CI=95%[79]]
- Haplogroup M9a1a – Han (Hebei, Henan, Shaanxi, Anhui, Zhejiang, Hunan, Yunnan, Guangdong, Hong Kong, Taiwan), Manchu (Jilin), Korean (South Korea), Hui (Qinghai), Kazakh (Ili), Kyrgyz (Kyrgyzstan), Nepal [TMRCA 16,500 (95% CI 12,800 <-> 20,900) ybp[61]]
- Haplogroup M9a4
- Haplogroup M9a4a – Kinh (Hanoi), Han (Shaanxi, Shandong, Zhejiang, Taiwan, Sichuan, Guangdong), Li (Hainan), Mulam (Guangxi), Jino (Xishuangbanna), Dai (Xishuangbanna), Chiang Mai
- Haplogroup M9a4b – Kinh (Hanoi), South Korea
- Haplogroup M9a5 – Han (Hunan, Hong Kong), Thailand, Pubiao (Malipo), Li (Hainan), Mulam (Luocheng), Zhuang (Bama Yao Autonomous County), Kinh (Hanoi)
- Haplogroup M9a1 – Han (Hunan) [TMRCA 19,500 (95% CI 13,800 <-> 26,700) ybp[61]]
- Haplogroup M9b – Han (Luocheng, Dujiangyan, Shaanxi), Cham (Binh Thuan), Mulam (Luocheng), Bouyei (Guizhou), Yi (Hezhang), Bunu (Dahua), Hui (Ili), Thailand (Phuan from Sukhothai Province[58][61])
- Haplogroup M9a – Han (Guangdong, Guangxi, Yunnan, Sichuan, Hunan, Taiwan, Anhui, Shaanxi, Shandong, Hebei), Korean (South Korea), Tujia (Hunan), Kinh (Hue), Mongol (Hohhot), Japanese, Lhoba[61] [TMRCA 23,000 (95% CI 18,100 <-> 28,800) ybp[61]]
- Haplogroup E – a subclade of M9 – found especially in Taiwan (Aboriginal peoples), Maritime Southeast Asia, and the Mariana Islands [TMRCA 23,695.4 ± 6,902.4 ybp; CI=95%[79]]
- Haplogroup M9a'b
- Haplogroup M10 [16] – small clade found in East Asia, Southeast Asia, Bangladesh, Central Asia, Saudi Arabia, southern Siberia, Russia, Belarus, and Poland [TMRCA 23,600 (95% CI 17,100 <-> 31,700) ybp[61]]
- M10-514C!/A15218G/C16362T!
- M10-514C!/A15218G/C16362T!* – Poland
- M10-T3167C/C4140T/T8793C/C12549T/A13152G/T14502C/C15040T/T15071C
- M10a [TMRCA 16,700 (95% CI 11,800 <-> 22,800) ybp[61]]
- M10a* – Myanmar
- M10a1 – China, Thailand, Myanmar, Japan, Shor, Daur [TMRCA 14,400 (95% CI 11,100 <-> 18,400) ybp[61]]
- M10a1* – Myanmar, Central Thailand (Mon), Japan (Aichi)
- M10a1-G16129A!!!
- M10a1-A13105G!/T16362C – Shor, Daur
- M10a1a
- M10a1a* – China, Saudi Arabia
- M10a1a1
- M10a1a1a – Mongolia,[80] Korea, Japan, China (Han from Kunming, etc.), Iron Age Black Sea Scythian[81]
- M10a1a1b – Altai, Korea, Japan, China
- M10a1a1b* – Chinese Uyghur
- M10a1a1b1
- M10a1a1b1* – Japan, China
- M10a1a1b1a – China
- M10a1a1b1b – China
- M10a1a1b1c – Uyghur, Altai-Kizhi[80]
- M10a1a1b2
- M10a1a1b2* – Taiwan (Hakka)
- M10a1a1b2a – Japan
- M10a1a2 – Northern Thailand (Khon Mueang from Lamphun Province[58])
- M10a1a3 – Taiwan
- M10a1b
- M10a1b* – South Korea, China (Han from Tai'an, etc.), India (Gallong,[67] etc.)
- M10a1b1 – China (Makatao)
- M10a1b2 – China (Tingri County)
- M10a1c – China
- M10a2 – Japan (Aichi), Kalmyk, Russian (northwestern Russia)
- M10a [TMRCA 16,700 (95% CI 11,800 <-> 22,800) ybp[61]]
- M10b – China (Shui[61]), Vietnam (Cờ Lao)[61]
- M10-514C!/A15218G/C16362T!
- Haplogroup M11 [17] – small clade found especially among the Chinese and also in some Japanese, Koreans, Oroqen, Yi, Tibetans, Tajiks in Dushanbe, Tajikistan,[50] and Bangladeshis[31] [TMRCA 20,987.7 ± 5,740.8 ybp; CI=95%[79]]
- Haplogroup M11* – Myanmar
- Haplogroup M11a'b'd
- Haplogroup M11a'b [TMRCA 16,209.0 ± 4,396.8 ybp; CI=95%[79]]
- Haplogroup M11a – Korea, Turkey [TMRCA 11,972.3 ± 3,523.2 ybp; CI=95%[79]]
- Haplogroup M11b [TMRCA 12,962.0 ± 4,819.2 ybp; CI=95%[79]]
- Haplogroup M11b1 – Taiwan (Minnan)
- Haplogroup M11b1a – Japan
- Haplogroup M11b1a1 – Japan, Han (Tai'an)
- Haplogroup M11b1a – Japan
- Haplogroup M11b2 – Japanese (Hokkaido), China, Altai-Kizhi, Tajik (Dushanbe)
- Haplogroup M11b1 – Taiwan (Minnan)
- Haplogroup M11d – China, Teleut, Kyrgyz, Iran
- Haplogroup M11a'b [TMRCA 16,209.0 ± 4,396.8 ybp; CI=95%[79]]
- Haplogroup M11c – Japan, Korea
- Haplogroup M12'G
- Haplogroup M12 [18] – small clade found especially among the aborigines of Hainan Island as well as in other populations of China,[34] Japan, Korea, Pashtuns, Tibet, Myanmar, Thailand, Cambodia, and Vietnam [TMRCA 31,287.5 ± 5,731.2 ybp; CI=95%[79]]
- Haplogroup M12a [TMRCA 26,020.6 ± 5,808.0 ybp; CI=95%[79]]
- Haplogroup M12a1 – Thailand/Laos
- Haplogroup M12a1a – Thailand (Htin in Phayao Province, Black Tai in Kanchanaburi Province, Mon in Kanchanaburi Province, Khon Mueang in Chiang Mai Province[58]), Laos (Lao in Luang Prabang[58]), Hainan
- Haplogroup M12a1a1 – China (esp. Hainan)
- Haplogroup M12a1a2 – Hainan, Tu
- Haplogroup M12a1b – Tibet, Thailand (Blang in Chiang Rai Province, Khon Mueang in Chiang Rai Province, Palaung in Chiang Mai Province, Mon in Kanchanaburi Province[58]), Hainan, Vietnam
- Haplogroup M12a1a – Thailand (Htin in Phayao Province, Black Tai in Kanchanaburi Province, Mon in Kanchanaburi Province, Khon Mueang in Chiang Mai Province[58]), Laos (Lao in Luang Prabang[58]), Hainan
- Haplogroup M12a2 – Thailand, Hainan, Myanmar
- Haplogroup M12a1 – Thailand/Laos
- Haplogroup M12b – Thailand (Khmu in Nan Province[58])
- Haplogroup M12b1 – Vietnam, Myanmar
- Haplogroup M12b1a – Laos (Lao in Vientiane[58])
- Haplogroup M12b1a1
- Haplogroup M12b1a2 – Thailand (Soa in Sakon Nakhon Province[58])
- Haplogroup M12b1a2a – Cambodia, Malaysia
- Haplogroup M12b1a2b – Cambodia
- Haplogroup M12b1b – Thailand (Suay in Surin Province, Khmer in Surin Province, Lao Isan in Roi Et Province, Black Tai in Loei Province[58]), Cambodia
- Haplogroup M12b1a – Laos (Lao in Vientiane[58])
- Haplogroup M12b2 – Thailand, Hainan
- Haplogroup M12b2a – Cambodia
- Haplogroup M12b1 – Vietnam, Myanmar
- Haplogroup M12a [TMRCA 26,020.6 ± 5,808.0 ybp; CI=95%[79]]
- Haplogroup G [19] – found especially in Japan, Mongolia, and Tibet and in indigenous peoples of Kamchatka (Koryaks, Alyutors, Itelmens), with some isolated instances in diverse places of Asia [TMRCA 31,614.8 ± 5,193.6 ybp; CI=95%[79]]
- Haplogroup G1 – Japan [TMRCA 21,492.9 ± 5,414.4 ybp; CI=95%[79]]
- Haplogroup G1a – China (Uyghurs), Thailand (Black Lahu in Mae Hong Son Province[62]) [TMRCA 18,139.1 ± 5,462.4 ybp; CI=95%[79]] (TMRCA 18,800 [95% CI 12,600 <-> 26,900] ybp[61])
- Haplogroup G1a1 – Korea, Vietnam (Dao), China (Sarikolis, Uyghurs, etc.), Tajikistan (Pamiris), Russia (Todzhin) [TMRCA 12,200 (95% CI 10,100 <-> 14,600) ybp][61]
- Haplogroup G1a1a – Japan, Korea,[38] Taiwan [TMRCA 5,200 (95% CI 3,800 <-> 7,000) ybp][61]
- Haplogroup G1a1a1 – Japan, Korea, China (Daur, Korean in Arun Banner) (TMRCA 3,100 [95% CI 1,250 <-> 6,300] ybp[61])
- Haplogroup G1a1a2 – Japan
- Haplogroup G1a1a3 – Japan
- Haplogroup G1a1a4 – Japan, Korea
- Haplogroup G1a1b – Taiwan (Makatao[82]), Russia (Altai-Kizhi[38])
- Haplogroup G1a1a – Japan, Korea,[38] Taiwan [TMRCA 5,200 (95% CI 3,800 <-> 7,000) ybp][61]
- Haplogroup G1a2'3
- Haplogroup G1a1 – Korea, Vietnam (Dao), China (Sarikolis, Uyghurs, etc.), Tajikistan (Pamiris), Russia (Todzhin) [TMRCA 12,200 (95% CI 10,100 <-> 14,600) ybp][61]
- Haplogroup G1b [TMRCA 7,246.3 ± 5,088.0 ybp; CI=95%[79]] (TMRCA 10,300 [95% CI 7,000 <-> 14,700] ybp[61])
- Haplogroup G1b1 – (TMRCA 8,400 [95% CI 6,200 <-> 11,100] ybp[61]) Koryaks (Magadan Oblast,[38] Severo-Evensk District of Magadan Oblast),[citation needed] Nivkhs,[73] Evens (Kamchatka[73]), Yakut (HGDP)
- Haplogroup G1b2'3'4 (G1b-G16129A!) – (TMRCA 7,700 [95% CI 5,000 <-> 11,400] ybp[61]) Evenks (Iengra[73]), Evens (Magadan region[38]), Orok, Nivkhs, Koryaks (Magadan Oblast,[38] Severo-Evensk District of Magadan Oblast,[citation needed] Kamchatka[73])
- Haplogroup G1b2 – (TMRCA 2,600 [95% CI 375 <-> 9,300] ybp[61]) Koryaks (Magadan region,[38] Severo-Evensk District of Magadan Region),[citation needed] Chukchi
- Haplogroup G1b3 – (TMRCA 4,700 [95% CI 1,000 <-> 13,500] ybp[61]) Chukchi (Anadyr),[38] Evens (Kamchatka)[73]
- Haplogroup G1b4 – (TMRCA 1,450 [95% CI 225 <-> 5,100] ybp[61]) Yukaghirs,[73] Even (Tompo[73])
- Haplogroup G1c – Korea (Seoul),[38] China (Han from Lanzhou,[74] etc.), Thailand (Black and Red Lahu in Mae Hong Son Province[62]), Malaysia (Seletar) [TMRCA 12,910.6 ± 6,009.6 ybp; CI=95%[79]] (TMRCA 17,200 [95% CI 11,600 <-> 24,600] ybp[61])
- Haplogroup G1c1 – Han (Sichuan, Tai'an)
- Haplogroup G1c2 – China (including at least one Han from Beijing)
- Haplogroup G1a – China (Uyghurs), Thailand (Black Lahu in Mae Hong Son Province[62]) [TMRCA 18,139.1 ± 5,462.4 ybp; CI=95%[79]] (TMRCA 18,800 [95% CI 12,600 <-> 26,900] ybp[61])
- Haplogroup G2 – Cambodia [TMRCA 26,787.5 ± 4,617.6 ybp; CI=95%[79]]
- Haplogroup G2a'c – China,[84] USA[84]
- Haplogroup G2a [TMRCA 17,145.5 ± 5,270.4 ybp; CI=95%[79]]
- Haplogroup G2a1 – Korea,[84] China[84][85] (Hebei,[86] Eastern China[87]), Tibetans (Chamdo,[88][61] Lhasa,[31] Tingri[89]), Uyghur (Artux[50]), Daur, India (Ladakh, Punjabi Hindu), Myanmar (Bamar from Kayin State[90]), Thailand, Singapore,[61] Kyrgyz, Kazakhstan,[84] Afghanistan (Balkh[61]), Israel (haMerkaz[61]), Saudi Arabia,[84] Russia[84] (Chelyabinsk Oblast,[61] Taimyr Evenk,[73] Tuvinian[91]), Belarus (Lipka Tatars[92]), Norway,[84] USA ("caucasian"[93]) [TMRCA 14,045.7 ± 4,742.4 ybp; CI=95%[79]]
- Haplogroup G2a1a (C12525T) - Georgia (Abkhaz[61]), Karachay (Karachay-Cherkess Republic[92][61]), Yakuts (Central[73][61]), Buryat (Inner Mongolia[92][61])
- Haplogroup G2a1b (A12753G)
- Haplogroup G2a1-T16189C! - China (Han from Yili,[95] Tibetan from Shigatse[31]), Korea,[84] Japan,[61][84] Vietnam (Nung[96][61]), Georgian
- Haplogroup G2a1-T16189C!-A16194G - Japan (Aichi[75][61])
- Haplogroup G2a1-T16189C!-C3654T - Eastern China[87]
- Haplogroup G2a1-T16189C!-G16526A - Uyghurs[61]
- Haplogroup G2a1-T16189C!-T15784C - Korea[61]
- Haplogroup G2a1d – Kyrgyz (Artux[50]), Uzbek (Uzbekistan[92]), Poland[84] [TMRCA 7,303.3 ± 3,657.6 ybp; CI=95%[79]]
- Haplogroup G2a1e – Japan,[61] Korea[61][84]
- Haplogroup G2a1f - China[98] (Taihang area in Henan province[100][61])
- Haplogroup G2a1g – China,[85][98][61] Mongols, Karakalpak
- Haplogroup G2a1-G13194A - Tibetan (Tingri[89][61])
- Haplogroup G2a1i - Lhoba[89]
- Haplogroup G2a1j (C5840T) - Thailand (Tai Yuan from North Thailand[94]), China
- Haplogroup G2a1k (A8521G) - Poland[102][61]
- Haplogroup G2a1l (C10043T) - Russia (Pskov Oblast[102]), Norway (Buskerud[61])
- Haplogroup G2a1m (C8766T) - Buryat (Inner Mongolia[92])
- Haplogroup G2a1n (T14180C) - Kyrgyzstan (Kyrgyz[92])
- Haplogroup G2a1o (T173C) - China (Henan Province[104])
- Haplogroup G2a1p (A374G) - Ladakh[61]
- Haplogroup G2a1q (T16356C) - Deng,[89] China (Fujian[61])
- Haplogroup G2a-T152C! - China (Eastern China[87]), Thai (Chanthaburi[61])
- Haplogroup G2a2 – China, Uyghur, Kyrgyz (Artux,[50] Tashkurgan[50]), Kazakhstan, Chelkan (Turochak, Biyka, Kurmach-Baigol), Nogai (Karachay-Cherkess Republic[92]), Yakut[64][61] (Vilyuy,[73] Northeast,[73] Central[73]), Buryat (Kizhinginsky District,[101] Olkhonsky District,[101] Tunkinsky District,[101] Bokhansky District[101]), Hazara (Pakistan[64][61]), Volga Tatar (Republic of Tatarstan[92]), Hungary, Finland
- Haplogroup G2a2a – China,[61] Uyghur,[61] Buryat (Bulagad from Bokhansky District[101]), Soyot,[101] Turkey, Bashkortostan,[61] Poland[61][105]
- Haplogroup G2a2b (G12007A) – Buryat (Tunkinsky District,[101][61] Ekhirit-Bulagatsky District[101][61])
- Haplogroup G2a2c (T16136C) – Uyghur[61]
- Haplogroup G2a2d (C5456T) - Uyghur[61]
- Haplogroup G2a2d1 (T8978C) - Uyghur[61]
- Haplogroup G2a2e (T4772C) - Buryat (Ekhirid from Kurumkansky District,[101][61] Bulagad/Ekhirid from Ekhirit-Bulagatsky District[101][61])
- Haplogroup G2a-T152C!-C16262T - ancient DNA from Irkutsk Oblast (Irk067 from the Novyj Kachug site at the left bank of the upper Lena River, cal BC 3755 to cal BC 3640[106][61])
- Haplogroup G2a4 – China (Taihang area in Henan province[100][61]), Taiwan (Taitung[108][61]), Ukraine[105][61]
- Haplogroup G2a5 – Japan,[61] Korea,[61] Kyrgyz, Kazakh, Karakalpak, Telengit, Tubalar, Yakut,[72] Balkar (Kabardino-Balkaria[61]), Buryat (Selenginsky District[101][61])
- Haplogroup G2a-T152C!-T1189C - Uyghur[61]
- Haplogroup G2a-T152C!-C3351T - China (Jiangxi Province,[104] Eastern China[87]), Buryat (Ekhirid/Bulagad from Olkhonsky District[101])
- Haplogroup G2a-T152C!-C10834T - China (Jilin[61]), Thailand (Iu Mien from Nan Province[62])
- Haplogroup G2a2 – China, Uyghur, Kyrgyz (Artux,[50] Tashkurgan[50]), Kazakhstan, Chelkan (Turochak, Biyka, Kurmach-Baigol), Nogai (Karachay-Cherkess Republic[92]), Yakut[64][61] (Vilyuy,[73] Northeast,[73] Central[73]), Buryat (Kizhinginsky District,[101] Olkhonsky District,[101] Tunkinsky District,[101] Bokhansky District[101]), Hazara (Pakistan[64][61]), Volga Tatar (Republic of Tatarstan[92]), Hungary, Finland
- Haplogroup G2a1 – Korea,[84] China[84][85] (Hebei,[86] Eastern China[87]), Tibetans (Chamdo,[88][61] Lhasa,[31] Tingri[89]), Uyghur (Artux[50]), Daur, India (Ladakh, Punjabi Hindu), Myanmar (Bamar from Kayin State[90]), Thailand, Singapore,[61] Kyrgyz, Kazakhstan,[84] Afghanistan (Balkh[61]), Israel (haMerkaz[61]), Saudi Arabia,[84] Russia[84] (Chelyabinsk Oblast,[61] Taimyr Evenk,[73] Tuvinian[91]), Belarus (Lipka Tatars[92]), Norway,[84] USA ("caucasian"[93]) [TMRCA 14,045.7 ± 4,742.4 ybp; CI=95%[79]]
- Haplogroup G2-A3397G
- Haplogroup G2c – China[98] (Taihang area in Henan province[100]), Uyghurs[61]
- Haplogroup G2a [TMRCA 17,145.5 ± 5,270.4 ybp; CI=95%[79]]
- Haplogroup G2b [TMRCA 22,776.4 ± 5,059.2 ybp; CI=95%[79]]
- Haplogroup G2b1 – Tibetan (Shannan[31]), Cambodia[84] [TMRCA 16,830.4 ± 5,616.0 ybp; CI=95%[79]]
- Haplogroup G2b1a – China[98] (Chongqing,[61] Shaanxi[61]), Thailand (Nyaw from Nakhon Phanom Province[58]), Kyrgyzstan,[61] Spain[84] [TMRCA 13,366.8 ± 5,443.2 ybp; CI=95%[79]]
- Haplogroup G2b1a1 – Thailand (Tai Yuan from Uttaradit Province, Khon Mueang from Chiang Mai Province,[58] Karen from Mae Hong Son Province[94]), Myanmar (Karen from Kayin State[90] and Bago Division[90])
- Haplogroup G2b1a2 – Ewenki from Inner Mongolia[95]
- Haplogroup G2b1a3 (A4562T) - Taiwan (Hakka[82])
- Haplogroup G2b1a4 (C4599T) - Thailand (Tai Khuen from Northern Thailand,[94] Tai Lue from Chiang Rai Province[94])
- Haplogroup G2b1a5 (T11353C) - China (Hebei Province[111]), Dungan[61]
- Haplogroup G2b1b – Uyghurs,[61] Tibetan (Shannan[31]), India (Lachungpa[67]) [TMRCA 4,662.1 ± 4,492.8 ybp; CI=95%[79]]
- Haplogroup G2b1a – China[98] (Chongqing,[61] Shaanxi[61]), Thailand (Nyaw from Nakhon Phanom Province[58]), Kyrgyzstan,[61] Spain[84] [TMRCA 13,366.8 ± 5,443.2 ybp; CI=95%[79]]
- Haplogroup G2b2 – Late Medieval eastern Mongolia (SHU001[112][61]), Sweden,[84] Russia,[84] Scotland,[84] Iraq,[84] Japan[84] [TMRCA 20,476.0 ± 5,481.6 ybp; CI=95%[79]]
- Haplogroup G2b1 – Tibetan (Shannan[31]), Cambodia[84] [TMRCA 16,830.4 ± 5,616.0 ybp; CI=95%[79]]
- Haplogroup G2d (A11443G) - Thailand (Tai Yuan in Central Thailand,[94] Iu Mien in Phayao Province[62])
- Haplogroup G2a'c – China,[84] USA[84]
- Haplogroup G3 [TMRCA 23,919.9 ± 6,931.2 ybp; CI=95%[79]]
- Haplogroup G3a
- Haplogroup G3a1'2 – Pakistan (Azad Kashmir)
- Haplogroup G3a1 [TMRCA 7,300 (95% CI 3,700 <-> 13,000) ybp][61]
- Haplogroup G3a1a – Tibetan (Shigatse[113]), Sherpa,[114] Naxi,[64][61] Hani[115]
- Haplogroup G3a1b – Tibet[61] (Nagqu[113]), Sarikoli (Tashkurgan[50])
- Haplogroup G3a1c (C2389A) - skeletal remains of Bo people from Chang'an Township, Weixin County, Zhaotong, Yunnan, China and similar people from Yangxu Town, Youjiang District, Baise, Guangxi, China and Tham Lod rockshelter, Pang Mapha District, Mae Hong Son Province, Thailand[116]
- Haplogroup G3a1'2-A12661G - Uyghur[61]
- Haplogroup G3a2 [TMRCA 12,600 (95% CI 3,900 <-> 30,200) ybp][61]
- Haplogroup G3a1 [TMRCA 7,300 (95% CI 3,700 <-> 13,000) ybp][61]
- Haplogroup G3a3 [TMRCA 12,900 (95% CI 6,000 <-> 24,200) ybp][61] – China,[110] Buryat (Khori from Khorinsky District[101]), Ukraine,[84] Poland,[84] Slovakia,[84] Albania,[84] United Kingdom[84]
- Haplogroup G3a1'2 – Pakistan (Azad Kashmir)
- Haplogroup G3b [TMRCA 17,800 (95% CI 12,800 <-> 24,100) ybp][61] - India (Lachungpa[67]), Tibetan (Lhasa[31]), Lhoba[89]
- Haplogroup G3a
- Haplogroup G4 – Japan [TMRCA 7,500 (95% CI 3,100 <-> 15,500) ybp][61]
- Haplogroup G5 – Vietnam (Dao[96]), Thailand (Lao Isan from Nakhon Ratchasima Province[62]) [TMRCA 7,900 (95% CI 5,400 <-> 11,200) ybp][61]
- Haplogroup G1 – Japan [TMRCA 21,492.9 ± 5,414.4 ybp; CI=95%[79]]
- Haplogroup M12 [18] – small clade found especially among the aborigines of Hainan Island as well as in other populations of China,[34] Japan, Korea, Pashtuns, Tibet, Myanmar, Thailand, Cambodia, and Vietnam [TMRCA 31,287.5 ± 5,731.2 ybp; CI=95%[79]]
- Haplogroup M13'46'61
- Haplogroup M13 – small clade found among Tibetans in Tibet,[31] Oirat Mongols in Xinjiang,[120] Barghuts in Hulunbuir,[80] Koreans,[115] and Yakuts and Dolgans in central Siberia[121] [TMRCA 43,169.4 ± 4,944.0 ybp; CI=95%[79]]
- Haplogroup M46 – Myanmar, Moken, Urak Lawoi, Malagasy
- Haplogroup M61 – Thailand (Phuan in Suphan Buri Province, Phuan in Phichit Province, Phuan in Sukhothai Province, Saek in Nakhon Phanom Province[58]), Myanmar, Vietnam, Borneo
- Haplogroup M61a – China (Yi, Tibet, Lachungpa)
- Haplogroup M14 – Australia (Kalumburu), Saudi Arabia, India, Tibet?[31]
- Haplogroup M15 – Australia (Kalumburu), Tibet?[31]
- Haplogroup M17 – found in Luzon,[40] Chams,[41][122] Maniq,[122] Mon,[122] Blang,[122] Lawa,[122] Thai,[122] and Laotians[122]
- Haplogroup M17a – Thailand, Vietnam (Cham)
- Haplogroup M17c – Vietnam (Cham)
- Haplogroup M17c1 – Philippines (Abaknon)
- Haplogroup M17c1a – Indonesia
- Haplogroup M17c1a1 – Philippines (Abaknon)
- Haplogroup M17c1a1a – Cambodia (incl. Stieng)
- Haplogroup M17c1a1 – Philippines (Abaknon)
- Haplogroup M17c1a – Indonesia
- Haplogroup M17c1 – Philippines (Abaknon)
- Haplogroup M19'53
- Haplogroup M19 – found in the Batak people of Palawan[123]
- Haplogroup M53 – India
- Haplogroup M53b – Kamar of Chhattisgarh
- Haplogroup M21 [20] – small clade found in SE Asia (Semang, Semelai, Temuan,[122] Jehai,[122] Thailand, Maniq,[122] Mon,[122] Karen,[122] Indonesia), China, and Bangladesh
- Haplogroup M21a – Batek
- Haplogroup M21b
- Haplogroup M21b1 – Cambodia (Lao, Tompoun)
- Haplogroup M21b1a – Semelai, Philippines (Cuyunon of Palawan Island), Indonesia
- Haplogroup M21b2 – Moken, Cham, Malaysia, India
- Haplogroup M21b1 – Cambodia (Lao, Tompoun)
- Haplogroup M22 – Drung (Yunnan)
- Haplogroup M22a – Vietnam (Kinh, Cham), Temuan
- Haplogroup M22b – Vietnam (Kinh), China (Han in Meizhou), Cambodia
- Haplogroup M23'75 – China
- M23 – found in Madagascar , South Africa, USA, Canada
- M75 – found in China,[59] USA
- Haplogroup M24'41 [124]
- Haplogroup M24
- Haplogroup M24a – found in Thailand, Cambodia (Khmer), Palawan (Tagbanua),[123] and China
- Haplogroup M24b – found in Thailand, Cambodia (Jarai), and China
- Haplogroup M41 – found in South Asia
- Haplogroup M24
- Haplogroup M27 [21] – found in Melanesia
- Haplogroup M28 [22] – found in Melanesia and in a single Han individual from China[31]
- Haplogroup M29'Q
- Haplogroup M29 [23] – found in Melanesia
- Haplogroup Q [24] – found in Melanesia and Australia (Aboriginal peoples)
- Haplogroup M31 [25] – found among the Onge, in the Andaman Islands[21]
- Haplogroup M31a
- Haplogroup M31a1
- Haplogroup M31a1a – Andaman Islands
- Haplogroup M31a1b – Andaman Islands
- Haplogroup M31a2
- Haplogroup M31a2a – northern India (Munda, etc.), Myanmar
- Haplogroup M31a2b – India (Paudibhuiya)
- Haplogroup M31a1
- Haplogroup M31b'c
- Haplogroup M31b – northern India, Nepal, Myanmar
- Haplogroup M31c – Nepal (Tharu)
- Haplogroup M31a
- Haplogroup M32'56 – Thailand
- Haplogroup M33 – India
- Haplogroup M33a – found in Nepal (Tharu),[65][50] India
- Haplogroup M33a1
- Haplogroup M33a1a – Lepcha, Tharu
- Haplogroup M33a1b – Dongri Bhil, Gujarat[21]
- Haplogroup M33a2'3
- Haplogroup M33a1
- Haplogroup M33b'c (M33-T16362C, M33-a) – northern India[125]
- Haplogroup M33b – Nepal (Tharu), India
- Haplogroup M33b1 – Sonowal Kachari, Dai (Jianshui)
- Haplogroup M33b2 – India, Nepal (Kathmandu)
- Haplogroup M33c – Ashkenazi Jews (Lithuania, Russia, Belarus, Ukraine, Romania, Hungary, Austria, Latvia, Poland),[126][127] Han Chinese[126] (Guangdong, Shaanxi, Sichuan, Hunan, Jiangsu, Jilin), Yao (Jianghua, Guangxi, Guangdong),[126] Miao (Hunan),[126] Tibetans (Yunnan),[126] Zhuang (Guangxi),[126] speakers of Kam-Tai languages (Guizhou),[126] Thai (Thailand),[126] Vietnamese[126]
- Haplogroup M33b – Nepal (Tharu), India
- Haplogroup M33d – India (Malpaharia, etc.)
- Haplogroup M33a – found in Nepal (Tharu),[65][50] India
- Haplogroup M34'57
- Haplogroup M34 [27] – small clade found in South Asia
- Haplogroup M34a – found in Karnataka, India[21]
- Haplogroup M34a1 – India
- Haplogroup M34a1a – India, Myanmar
- Haplogroup M34a2 – India (Pauri Bhuiya, Munda)
- Haplogroup M34a1 – India
- Haplogroup M34b – India (Nihal, etc.)
- Haplogroup M34a – found in Karnataka, India[21]
- Haplogroup M57 – India (Kathakur)
- Haplogroup M57a – India (Katkari, Nihal), England, Ireland
- Haplogroup M57b – India (Kathakur)
- Haplogroup M57b1 – India (Dongri Bhil)
- Haplogroup M34 [27] – small clade found in South Asia
- Haplogroup M35 [28] – Nepal (Tharu)
- Haplogroup M35a – Sarikoli,[50] Armenia, Mesolithic Sri Lanka[66]
- Haplogroup M35a1 – India (Andh, Dongri Bhil), Mauritius
- Haplogroup M35a1a – India (Betta Kurumba, Mullukurunan)
- Haplogroup M35a2 – India (Andh, etc.)
- Haplogroup M35a1 – India (Andh, Dongri Bhil), Mauritius
- Haplogroup M35b – found in Karnataka, Ladakh, Nepal (Tharu), Myanmar, Thailand, Slovakia[128]
- Haplogroup M35b1'2'3 – India, Germany, USA
- Haplogroup M35b1 – India (Madia)
- Haplogroup M35b2 – India (Kathodi, Munda), Russia
- Haplogroup M35b3 – India (Ladakh)
- Haplogroup M35b4 – India (Toto), Nepal (Kathmandu)
- Haplogroup M35b1'2'3 – India, Germany, USA
- Haplogroup M35c – India (Kathodi, Andh)
- Haplogroup M35a – Sarikoli,[50] Armenia, Mesolithic Sri Lanka[66]
- Haplogroup M39'70 [129]
- Haplogroup M40 [30] – found in South Asia[21]
- Haplogroup M40a – Yemen
- Haplogroup M40a1
- Haplogroup M40a1a
- Haplogroup M40a1b
- Haplogroup M40a1
- Haplogroup M40a – Yemen
- Haplogroup M42'74 [130]
- Haplogroup M48 [32] – rare clade found in Saudi Arabia
- Haplogroup M49 – found among ancient specimens in the Euphrates valley
- Haplogroup M55'77 [132]
- Haplogroup M55 - Myanmar, Thailand, Malay
- Haplogroup M77 - Indonesia [133]
- Haplogroup M62'68 [134]
- Haplogroup M62
- Haplogroup M68 [135]
- Haplogroup M68a1
- Haplogroup M68a1a - Cambodia, Vietnam
- Haplogroup M68a1b
- Haplogroup M68a2
- Haplogroup M68a2a- Cambodia
- Haplogroup M68a2b
- Haplogroup M68a2c
- Haplogroup M68a1
- Haplogroup M71 - India
- Haplogroup M71a'b (M71-C151T) - India, Myanmar, Cambodia (Mel), Laos (Lao in Vientiane), Thailand (Lawa, Karen, Shan, Blang, Phuan, Lao Isan, Khon Mueang), Vietnam (Ede)
- Haplogroup M71a - Thailand (Thai, Tai Yuan, Khon Mueang), China (Fujian)
- Haplogroup M71a1 - China (Han from Xiamen and Lanzhou,[74] Naxi)
- Haplogroup M71a1a - China (Bouyei from Pingtang, etc.), Vietnam (Mang, Kinh), Thailand (Phuan from Central Thailand)
- Haplogroup M71a2 - India, Myanmar, Thailand (Thai from Central Thailand and Eastern Thailand,[136] Tai Yuan and Tai Khün from Northern Thailand,[136] Blang[58]), Indonesia, Philippines [40]
- Haplogroup M71a1 - China (Han from Xiamen and Lanzhou,[74] Naxi)
- Haplogroup M71b - Thailand (Thai from Western Thailand,[122] Khon Mueang from Chiang Mai Province,[58] Tai Yuan,[122] Tai Khün[122]), China (Bouyei from Pingtang,[59] Han from Dongguan[59])
- Haplogroup M71a - Thailand (Thai, Tai Yuan, Khon Mueang), China (Fujian)
- Haplogroup M71c - Thailand (Urak Lawoi, Moken, Thai from Central Thailand), Vietnam (Cham from Bình Thuận, Kinh)
- Haplogroup M71a'b (M71-C151T) - India, Myanmar, Cambodia (Mel), Laos (Lao in Vientiane), Thailand (Lawa, Karen, Shan, Blang, Phuan, Lao Isan, Khon Mueang), Vietnam (Ede)
- Haplogroup M73'79 [137]
- Haplogroup M73 [138]
- Haplogroup M73a
- Haplogroup M73a1
- Haplogroup M73a1a
- Haplogroup M73a1b - Vietnam
- Haplogroup M73a2 - Papua New Guinea, East Timor
- Haplogroup M73a3 - Philippines (Aklan)
- Haplogroup M73a1
- Haplogroup M73b - Indonesia
- Haplogroup M73b1 - Vietnam, Cambodia, Indonesia
- Haplogroup M73c
- Haplogroup M73a
- Haplogroup M79 - China [139]
- Haplogroup M73 [138]
- Haplogroup M80'D
- Haplogroup M80 – found in Batak people of Palawan[123]
- Haplogroup D – found in Eastern Eurasia, Native Americans, Central Asia[140] and occasionally also in West Asia and Europe.
Subclades
Tree
This phylogenetic tree of haplogroup M subclades is based on the paper by Mannis van Oven and Manfred Kayser Updated comprehensive phylogenetic tree of global human mitochondrial DNA variation[13] and subsequent published research.
- M
- M1
- M1a
- M1a1
- M1a1a
- M1a1b
- M1a1b1
- M1a1c
- M1a1d
- M1a1e
- M1a1f
- M1a2
- M1a2a
- M1a2b
- M1a3
- M1a3a
- M1a3b
- M1a4
- M1a5
- M1a1
- M1b
- M1b1
- M1b1a
- M1b2
- M1b2a
- M1b1
- M1a
- M2
- M2a
- M2a1
- M2a2
- M2a3
- M2b
- M2b1
- M2b2
- M2a
- M3
- M3a
- M4"45
- M4
- M4a
- M4b
- M4b1
- M18'38
- M18
- M38
- M30
- M30a
- M30b
- M30c
- M30c1
- M30c1a
- M30c1a1
- M30c1a
- M30c1
- M30d
- M37
- M37a
- M43
- M45
- M4
- M5
- M5a
- M5a1
- M5a1a
- M5a1b
- M5a2
- M5a2a
- M5a1
- M5a
- M6
- M7
- M7a
- M7a1
- M7a1a
- M7a1a1
- M7a1a1a
- M7a1a2
- M7a1a3
- M7a1a4
- M7a1a4a
- M7a1a5
- M7a1a6
- M7a1a7
- M7a1a1
- M7a1b
- M7a1a
- M7a2
- M7a2a
- M7a2b
- M7a1
- M7b'c'd'e
- M7b'd
- M7b
- M7b1'2
- M7b1
- M7b2
- M7b2a
- M7b2b
- M7b2c
- M7b3
- M7b3a
- M7b1'2
- M7d
- M7b
- M7c'e
- M7c
- M7c1
- M7c1a
- M7c1b
- M7c1b1
- M7c2
- M7c2a
- M7c3
- M7c3a
- M7c3b
- M7c3c
- M7c1
- M7e
- M7c
- M7b'd
- M7a
- M8
- M9
- M9a'b'c'd
- M9a'c'd
- M9a'd
- M9a
- M9a1
- M9a2
- M9a3
- M9d
- M9a
- M9c
- M9a'd
- M9b
- M9a'c'd
- E
- M9a'b'c'd
- M10'42
- M10
- M10a
- M10a1
- M10a2
- M10a
- M42
- M42a
- M10
- M11
- M11a
- M11b
- M12'G
- M12
- M12a
- G
- M12
- M13
- M13a
- M13a1
- M13a
- M14
- M15
- M21
- M21a'b
- M21a
- M21b
- M21c'd
- M21c
- M21d
- M21a'b
- M22
- M23
- M25
- M27
- M27a
- M27b
- M27c
- M28
- M28a
- M28b
- M29'Q
- M29
- M29a
- M29b
- Q
- M29
- M31
- M31a
- M31a1
- M31a1a
- M31a1b
- M31a2
- M31a1
- M31b'c
- M31b
- M31b1
- M31b2
- M31c
- M31b
- M31a
- M32'56
- M32
- M32a
- M32c
- M56
- M32
- M33
- M33a
- M33b
- M33c
- M34
- M34a
- M35
- M35a
- M35b
- M36
- M36a
- M39
- M39a
- M40
- M40a
- M41
- M44'52
- M44
- M52
- M46
- M47'50
- M47
- M50
- M48
- M49
- M51
- M52’58
- M52
- D
- M1
See also
- Genealogical DNA test
- Genetic genealogy
- Human mitochondrial genetics
- Population genetics
- Human mitochondrial DNA haplogroups
|
Phylogenetic tree of human mitochondrial DNA (mtDNA) haplogroups | |||||||||||||||||||||||||||||||||||||||
| Mitochondrial Eve (L) | |||||||||||||||||||||||||||||||||||||||
| L0 | L1–6 | ||||||||||||||||||||||||||||||||||||||
| L1 | L2 | L3 | L4 | L5 | L6 | ||||||||||||||||||||||||||||||||||
| M | N | ||||||||||||||||||||||||||||||||||||||
| CZ | D | E | G | Q | O | A | S | R | I | W | X | Y | |||||||||||||||||||||||||||
| C | Z | B | F | R0 | pre-JT | P | U | ||||||||||||||||||||||||||||||||
| HV | JT | K | |||||||||||||||||||||||||||||||||||||
| H | V | J | T | ||||||||||||||||||||||||||||||||||||
References
- ↑ 1.0 1.1 1.2 Soares, P; Alshamali, F; Pereira, J. B; Fernandes, V; Silva, N. M; Afonso, C; Costa, M. D; Musilova, E et al. (2011). "The Expansion of mtDNA Haplogroup L3 within and out of Africa". Molecular Biology and Evolution 29 (3): 915–927. doi:10.1093/molbev/msr245. PMID 22096215.
- ↑ 2.0 2.1 "Ancestral mitochondrial N lineage from the Neolithic 'green' Sahara". Sci Rep 9 (1): 3530. March 2019. doi:10.1038/s41598-019-39802-1. PMID 30837540. Bibcode: 2019NatSR...9.3530V.
- ↑ 3.0 3.1 3.2 3.3 3.4 3.5 3.6 3.7 Rajkumar (2005). "Phylogeny and antiquity of M macrohaplogroup inferred from complete mt DNA sequence of Indian specific lineages". BMC Evolutionary Biology 5: 26. doi:10.1186/1471-2148-5-26. PMID 15804362.
- ↑ 4.0 4.1 Macaulay, V; Hill, C; Achilli, A; Rengo, C; Clarke, D; Meehan, W; Blackburn, J; Semino, O et al. (2005). "Single, Rapid Coastal Settlement of Asia Revealed by Analysis of Complete Mitochondrial Genomes". Science 308 (5724): 1034–6. doi:10.1126/science.1109792. PMID 15890885. Bibcode: 2005Sci...308.1034M. http://psasir.upm.edu.my/id/eprint/40255/1/Single%2C%20rapid%20coastal%20settlement%20of%20Asia%20revealed%20by%20analysis%20of%20complete%20mitochondrial%20genomes.pdf.: "Haplogroup L3 (the African clade that gave rise to the two basal non-African clades, haplogroups M and N) is 84,000 years old, and haplogroups M and N themselves are almost identical in age at 63,000 years old, with haplogroup R diverging rapidly within haplogroup N 60,000 years ago."
- ↑ 5.00 5.01 5.02 5.03 5.04 5.05 5.06 5.07 5.08 5.09 5.10 Gonzalez (2007). "Mitochondrial lineage M1 traces an early human backflow to Africa". BMC Genomics 8: 223. doi:10.1186/1471-2164-8-223. PMID 17620140.
- ↑ 6.0 6.1 Abu-Amero, KK et al. (2008). "Mitochondrial DNA structure in the Arabian Peninsula". BMC Evolutionary Biology 8: 45. doi:10.1186/1471-2148-8-45. PMID 18269758.
- ↑ Kivisild, M; Kivisild, T; Metspalu, E; Parik, J; Hudjashov, G; Kaldma, K; Serk, P; Karmin, M et al. (2004). "Most of the extant mtDNA boundaries in South and Southwest Asia were likely shaped during the initial settlement of Eurasia by anatomically modern humans". BMC Genetics 5: 26. doi:10.1186/1471-2156-5-26. PMID 15339343.
- ↑ 8.0 8.1 8.2 Kivisild, T; Rootsi, S; Metspalu, M; Mastana, S; Kaldma, K; Parik, J; Metspalu, E; Adojaan, M et al. (2003). "The Genetic Heritage of the Earliest Settlers Persists Both in Indian Tribal and Caste Populations". American Journal of Human Genetics 72 (2): 313–32. doi:10.1086/346068. PMID 12536373.
- ↑ Marrero, Patricia; Abu-Amero, Khaled K.; Larruga, Jose M.; Cabrera, Vicente M. (2016). "Carriers of human mitochondrial DNA macrohaplogroup M colonized India from southeastern Asia". BMC Evolutionary Biology 16 (1): 246. doi:10.1186/s12862-016-0816-8. PMID 27832758.
- ↑ Quintana-Murci, Lluís (1999). "Where West Meets East: The Complex mtDNA Landscape of the Southwest and Central Asian Corridor". Am. J. Hum. Genet. 74 (5): 827–45. doi:10.1086/383236. PMID 15077202.
- ↑ Chandrasekar, Adimoolam; Kumar, Satish; Sreenath, Jwalapuram; Sarkar, Bishwa Nath; Urade, Bhaskar Pralhad; Mallick, Sujit; Bandopadhyay, Syam Sundar; Barua, Pinuma et al. (2009). "Updating Phylogeny of Mitochondrial DNA Macrohaplogroup M in India: Dispersal of Modern Human in South Asian Corridor". PLOS ONE 4 (10): e7447. doi:10.1371/journal.pone.0007447. PMID 19823670. Bibcode: 2009PLoSO...4.7447C.
- ↑ Osman, Maha M.. "Mitochondrial HVRI and whole mitogenome sequence variations portray similar scenarios on the genetic structure and ancestry of northeast Africans". Meta Gene. http://81.95.108.158/return-files/Mitochondrial%20HVRI.pdf.
- ↑ 13.0 13.1 van Oven, M et al. (2009). "Updated comprehensive phylogenetic tree of global human mitochondrial DNA variation". Human Mutation 30 (2): E386–E394. doi:10.1002/humu.20921. PMID 18853457.
- ↑ 14.0 14.1 14.2 14.3 14.4 14.5 14.6 14.7 Metspalu, M et al. (2004). "Most of the extant mtDNA boundaries in South and Southwest Asia were likely shaped during the initial settlement of Eurasia by anatomically modern humans". BMC Genetics 5: 26. doi:10.1186/1471-2156-5-26. PMID 15339343.
- ↑ Fregel, R; Cabrera, V; Larruga, JM; Abu-Amero, KK; González, AM (2015). "Carriers of Mitochondrial DNA Macrohaplogroup N Lineages Reached Australia around 50,000 Years Ago following a Northern Asian Route". PLOS ONE 10 (6): e0129839. doi:10.1371/journal.pone.0129839. PMID 26053380. Bibcode: 2015PLoSO..1029839F.
- ↑ Correcting for Purifying Selection: An Improved Human Mitochondrial Molecular Clock Supplementary material (p. 84). 2009. pp. 89. http://download.cell.com/AJHG/mmcs/journals/0002-9297/PIIS0002929709001633.mmc1.pdf.
- ↑ 17.0 17.1 17.2 Quintana (1999). Genetic evidence of an early exit of Homo sapiens sapiens from Africa through eastern Africa. http://users.clas.ufl.edu/krigbaum/proseminar/quintana-murci_naturegenetics_1999.pdf.
- ↑ Dubut, V et al. (2009). "Complete mitochondrial sequences for haplogroups M23 and M46: insights into the Asian ancestry of the Malagasy population". Human Biology 81 (4): 495–500. doi:10.3378/027.081.0407. PMID 20067372.
- ↑ Ricaut, FX et al. (2009). "A new deep branch of eurasian mtDNA macrohaplogroup M reveals additional complexity regarding the settlement of Madagascar". BMC Genomics 10: 605. doi:10.1186/1471-2164-10-605. PMID 20003445.
- ↑ 20.0 20.1 Maruyama, Sayaka; Minaguchi, Kiyoshi; Saitou, Naruya (2003). "Sequence polymorphisms of the mitochondrial DNA control region and phylogenetic analysis of mtDNA lineages in the Japanese population". Int J Legal Med 117 (4): 218–225. doi:10.1007/s00414-003-0379-2. PMID 12845447.
- ↑ 21.00 21.01 21.02 21.03 21.04 21.05 21.06 21.07 21.08 21.09 21.10 21.11 21.12 21.13 Thangaraj et al. (2006), In situ origin of deep rooting lineages of mitochondrial Macrohaplogroup 'M' in India, BMC Genomics 2006, 7:151
- ↑ Chandrasekar Adimoolam, Kumar SExpression error: Unrecognized word "etal". (2009). "Updating Phylogeny of Mitochondrial DNA Macrohaplogroup M in India: Dispersal of Modern Human in South Asian Corridor". PLOS ONE 4 (10): e7447. doi:10.1371/journal.pone.0007447. PMID 19823670. Bibcode: 2009PLoSO...4.7447C.
- ↑ 23.0 23.1 23.2 Olivieri et al. (2006), The mtDNA legacy of the Levantine early Upper Palaeolithic in Africa, Science. 2006 Dec 15;314(5806):1767-70
- ↑ Hudjashov, G; Kivisild, T; Underhill, PA; Endicott, P; Sanchez, JJ; Lin, AA; Shen, P; Oefner, P et al. (2007). "Revealing the prehistoric settlement of Australia by Y chromosome and mtDNA analysis". Proceedings of the National Academy of Sciences of the United States of America 104 (21): 8726–30. doi:10.1073/pnas.0702928104. PMID 17496137. Bibcode: 2007PNAS..104.8726H.
- ↑ Ghezzi (2005). "Mitochondrial DNA haplogroup K is associated with a lower risk of Parkinson's disease in Italians". European Journal of Human Genetics 13 (6): 748–752. doi:10.1038/sj.ejhg.5201425. PMID 15827561.
- ↑ Michael D. Petraglia; Bridget Allchin (2007), The evolution and history of human populations in South Asia, Springer, ISBN 978-1-4020-5561-4, https://books.google.com/books?id=Qm9GfjNlnRwC, "... As haplogroup M, except for the African sub-clade M1, is not notably present in regions west of the Indian subcontinent, while it covers the majority of Indian mtDNA variation ..."
- ↑ Endicott, Phillip; Metspalu, Mait; Stringer, Chris; Macaulay, Vincent; Cooper, Alan; Sanchez, Juan J. (2006-12-20). "Multiplexed SNP Typing of Ancient DNA Clarifies the Origin of Andaman mtDNA Haplogroups amongst South Asian Tribal Populations". PLOS ONE 1 (1): e81. doi:10.1371/journal.pone.0000081. PMID 17218991. Bibcode: 2006PLoSO...1...81E.
- ↑ The 200,000-Year Evolution of Homo sapiens sapiens Language and Myth Families based on the mtDNA Phylotree, Fossil mtDNA and Archaeology: A Thought Experiment (2014)[1]
- ↑ Hill, C.; Soares, P.; Mormina, M.; MacAulay, V.; Clarke, D.; Blumbach, P. B.; Vizuete-Forster, M.; Forster, P. et al. (2006). "A Mitochondrial Stratigraphy for Island Southeast Asia". American Journal of Human Genetics 80 (1): 29–43. doi:10.1086/510412. PMID 17160892.
- ↑ Hill, C; Soares, P; Mormina, M; Macaulay, V; Clarke, D; Blumbach, PB; Vizuete-Forster, M; Forster, P et al. (2007). "A mitochondrial stratigraphy for island southeast Asia". Am. J. Hum. Genet. 80 (1): 29–43. doi:10.1086/510412. PMID 17160892.
- ↑ 31.00 31.01 31.02 31.03 31.04 31.05 31.06 31.07 31.08 31.09 31.10 31.11 31.12 31.13 31.14 Ji, FuyunExpression error: Unrecognized word "etal". (2012). "Mitochondrial DNA variant associated with Leber hereditary optic neuropathy and high-altitude Tibetans". PNAS 109 (19): 7391–7396. doi:10.1073/pnas.1202484109. PMID 22517755. Bibcode: 2012PNAS..109.7391J.
- ↑ 32.0 32.1 32.2 32.3 32.4 32.5 Umetsu, KazuoExpression error: Unrecognized word "etal". (2005). "Multiplex amplified product-length polymorphism analysis of 36 mitochondrial single-nucleotide polymorphisms for haplogrouping of East Asian populations". Electrophoresis 26 (1): 91–98. doi:10.1002/elps.200406129. PMID 15624129.
- ↑ Asari, MasaruExpression error: Unrecognized word "etal". (2007). "Utility of haplogroup determination for forensic mtDNA analysis in the Japanese population". Legal Medicine 9 (5): 237–240. doi:10.1016/j.legalmed.2007.01.007. PMID 17467322.
- ↑ 34.0 34.1 34.2 Zheng, H-XExpression error: Unrecognized word "etal". (2011). "Major Population Expansion of East Asians Began before Neolithic Time: Evidence of mtDNA Genomes". PLOS ONE 6 (10): e25835. doi:10.1371/journal.pone.0025835. PMID 21998705. Bibcode: 2011PLoSO...625835Z.
- ↑ Edwin (2002). "Mitochondrial DNA diversity among five tribal populations of southern India". Current Science 83. http://www.iisc.ernet.in/currsci/jul252002/158.pdf. Retrieved 2008-10-25.
- ↑ Kim, WExpression error: Unrecognized word "etal". (2008). "Mitochondrial DNA Haplogroup Analysis Reveals no Association between the Common Genetic Lineages and Prostate Cancer in the Korean Population". PLOS ONE 3 (5): e2211. doi:10.1371/journal.pone.0002211. PMID 18493608. Bibcode: 2008PLoSO...3.2211K.
- ↑ 37.0 37.1 37.2 Jin, H-J; Tyler-Smith, C; Kim, W (2009). "The Peopling of Korea Revealed by Analyses of Mitochondrial DNA and Y-Chromosomal Markers". PLOS ONE 4 (1): e4210. doi:10.1371/journal.pone.0004210. PMID 19148289. Bibcode: 2009PLoSO...4.4210J.
- ↑ 38.00 38.01 38.02 38.03 38.04 38.05 38.06 38.07 38.08 38.09 38.10 Derenko, MiroslavaExpression error: Unrecognized word "etal". (2007). "Phylogeographic Analysis of Mitochondrial DNA in Northern Asian Populations". Am. J. Hum. Genet. 81 (5): 1025–1041. doi:10.1086/522933. PMID 17924343.
- ↑ Beom Hong, Seung; Cheol Kim, Ki; Kim, Wook (2014). "Mitochondrial DNA haplogroups and homogeneity in the Korean population". Genes & Genomics 36 (5): 583–590. doi:10.1007/s13258-014-0194-9.
- ↑ 40.0 40.1 40.2 Tabbada, Kristina A.Expression error: Unrecognized word "etal". (Jan 2010). "Philippine Mitochondrial DNA Diversity: A Populated Viaduct between Taiwan and Indonesia?". Mol. Biol. Evol. 27 (1): 21–31. doi:10.1093/molbev/msp215. PMID 19755666.
- ↑ 41.0 41.1 Peng, Min-ShengExpression error: Unrecognized word "etal". (2010). "Tracing the Austronesian Footprint in Mainland Southeast Asia: A Perspective from Mitochondrial DNA". Mol. Biol. Evol. 27 (10): 2417–2430. doi:10.1093/molbev/msq131. PMID 20513740.
- ↑ 42.0 42.1 Bodner, MartinExpression error: Unrecognized word "etal". (2011). "Southeast Asian diversity: first insights into the complex mtDNA structure of Laos". BMC Evolutionary Biology 11: 49. doi:10.1186/1471-2148-11-49. PMID 21333001.
- ↑ Kayser, ManfredExpression error: Unrecognized word "etal". (July 2008). "", (2008) "The Impact of the Austronesian Expansion: Evidence from mtDNA and Y Chromosome Diversity in the Admiralty Islands of Melanesia". Molecular Biology and Evolution 25 (7): 1362–1374. doi:10.1093/molbev/msn078. PMID 18390477.
- ↑ Hudjashov, GeorgiExpression error: Unrecognized word "etal". (2007). "Revealing the prehistoric settlement of Australia by Y chromosome and mtDNA analysis". PNAS 104 (21): 8726–8730. doi:10.1073/pnas.0702928104. PMID 17496137. Bibcode: 2007PNAS..104.8726H.
- ↑ Shipley, G. P.; Taylor, D. A.; Tyagi, A.; Tiwari, G.; Redd, A. (2015). "Genetic structure among Fijian island populations". Journal of Human Genetics 60 (2): 69–75. doi:10.1038/jhg.2014.105. PMID 25566758.
- ↑ Non, Amy. "ANALYSES OF GENETIC DATA WITHIN AN INTERDISCIPLINARY FRAMEWORK TO INVESTIGATE RECENT HUMAN EVOLUTIONARY HISTORY AND COMPLEX DISEASE". University of Florida. http://etd.fcla.edu/UF/UFE0041981/non_a.pdf.
- ↑ Holden. "MtDNA variation in North, East, and Central African populations gives clues to a possible back-migration from the Middle East". American Association of Physical Anthropologists. http://konig.la.utk.edu/AJPA_Suppl_40_web.htm.
- ↑ Luísa Pereira; Viktor Černý; María Cerezo; Nuno M Silva; Martin Hájek; Alžběta Vašíková; Martina Kujanová; Radim Brdička et al. (17 March 2010). "Linking the sub-Saharan and West Eurasian gene pools: maternal and paternal heritage of the Tuareg nomads from the African Sahel". European Journal of Human Genetics 18 (8): 915–923. doi:10.1038/ejhg.2010.21. PMID 20234393.
- ↑ 49.0 49.1 49.2 49.3 49.4 Derenko, MExpression error: Unrecognized word "etal". (2013). "Complete Mitochondrial DNA Diversity in Iranians". PLOS ONE 8 (11): e80673. doi:10.1371/journal.pone.0080673. PMID 24244704. Bibcode: 2013PLoSO...880673D.
- ↑ 50.00 50.01 50.02 50.03 50.04 50.05 50.06 50.07 50.08 50.09 50.10 50.11 50.12 50.13 50.14 50.15 50.16 50.17 50.18 50.19 50.20 50.21 Min-Sheng Peng, Weifang Xu, Jiao-Jiao Song, et al. (2017), "Mitochondrial genomes uncover the maternal history of the Pamir populations." European Journal of Human Genetics https://doi.org/10.1038/s41431-017-0028-8
- ↑ "mtDNA from the Early Bronze Age to the Roman Period Suggests a Genetic Link between the Indian Subcontinent and Mesopotamian Cradle of Civilization". PLOS ONE 8 (9): e73682. 2013. doi:10.1371/journal.pone.0073682. PMID 24040024. Bibcode: 2013PLoSO...873682W.
- ↑ Cosimo Posth; Gabriel Renaud; Alissa Mittnik; Dorothée G. Drucker; Hélène Rougier; Christophe Cupillard; Frédérique Valentin; Corinne Thevenet et al. (March 21, 2016). "Pleistocene Mitochondrial Genomes Suggest a Single Major Dispersal of Non-Africans and a Late Glacial Population Turnover in Europe". Current Biology 26 (6): 827–833. doi:10.1016/j.cub.2016.01.037. PMID 26853362.
- ↑ van de Loosdrecht (2018-03-15). "Pleistocene North African genomes link Near Eastern and sub-Saharan African human populations". Science 360 (6388): 548–552. doi:10.1126/science.aar8380. ISSN 0036-8075. PMID 29545507. Bibcode: 2018Sci...360..548V.
- ↑ Reyhan Yaka. "Archaeogenetics of Late Iron Age Çemialo Sırtı, Batman: Investigating maternal genetic continuity in North Mesopotamia since the Neolithic". bioRxiv 10.1101/172890.
- ↑ Schuenemann, Verena J. (2017). "Ancient Egyptian mummy genomes suggest an increase of Sub-Saharan African ancestry in post-Roman periods". Nature Communications 8: 15694. doi:10.1038/ncomms15694. PMID 28556824. Bibcode: 2017NatCo...815694S.
- ↑ "Ancient genomes from North Africa evidence prehistoric migrations to the Maghreb from both the Levant and Europe". 2018. bioRxiv 10.1101/191569.
- ↑ Konstantina Drosou; Campbell Price; Terence A. Brown (February 2018). "The kinship of two 12th Dynasty mummies revealed by ancient DNA sequencing". Journal of Archaeological Science 17: 793–797. doi:10.1016/j.jasrep.2017.12.025. https://www.research.manchester.ac.uk/portal/en/publications/the-kinship-of-two-12th-dynasty-mummies-revealed-by-ancient-dna-sequencing(6f7e0e83-90eb-4950-b451-cec627c5f6a4).html.
- ↑ 58.00 58.01 58.02 58.03 58.04 58.05 58.06 58.07 58.08 58.09 58.10 58.11 58.12 58.13 58.14 58.15 58.16 58.17 58.18 58.19 58.20 58.21 58.22 58.23 58.24 58.25 58.26 58.27 58.28 Kutanan,W., Kampuansai,J., Srikummool,M., Kangwanpong,D., Ghirotto,S., Brunelli,A. and Stoneking,M., "Complete mitochondrial genomes of Thai and Lao populations indicate an ancient origin of Austroasiatic groups and demic diffusion in the spread of Tai-Kadai languages." Hum. Genet. (2016).
- ↑ 59.0 59.1 59.2 59.3 Kong, Qing-Peng et al 2010, Large-Scale mtDNA Screening Reveals a Surprising Matrilineal Complexity in East Asia and Its Implications to the Peopling of the Region
- ↑ Jinam, Timothy A.; Hong, Lih-Chun; Phipps, Maude E.; Stoneking, Mark; Ameen, Mahmood; Edo, Juli; Hugo; SNP Consortium, Pan-Asian et al. (2012). "Evolutionary History of Continental Southeast Asians: 'Early Train' Hypothesis Based on Genetic Analysis of Mitochondrial and Autosomal DNA Data". Mol. Biol. Evol. 29 (11): 3513–3527. doi:10.1093/molbev/mss169. PMID 22729749.
- ↑ 61.000 61.001 61.002 61.003 61.004 61.005 61.006 61.007 61.008 61.009 61.010 61.011 61.012 61.013 61.014 61.015 61.016 61.017 61.018 61.019 61.020 61.021 61.022 61.023 61.024 61.025 61.026 61.027 61.028 61.029 61.030 61.031 61.032 61.033 61.034 61.035 61.036 61.037 61.038 61.039 61.040 61.041 61.042 61.043 61.044 61.045 61.046 61.047 61.048 61.049 61.050 61.051 61.052 61.053 61.054 61.055 61.056 61.057 61.058 61.059 61.060 61.061 61.062 61.063 61.064 61.065 61.066 61.067 61.068 61.069 61.070 61.071 61.072 61.073 61.074 61.075 61.076 61.077 61.078 61.079 61.080 61.081 61.082 61.083 61.084 61.085 61.086 61.087 61.088 61.089 61.090 61.091 61.092 61.093 61.094 61.095 61.096 61.097 61.098 61.099 61.100 61.101 61.102 61.103 61.104 61.105 61.106 61.107 61.108 61.109 61.110 61.111 61.112 61.113 61.114 61.115 61.116 61.117 61.118 61.119 61.120 61.121 61.122 61.123 61.124 61.125 61.126 61.127 61.128 61.129 61.130 61.131 61.132 61.133 YFull MTree 1.01.14609 as of August 21, 2019
- ↑ 62.00 62.01 62.02 62.03 62.04 62.05 62.06 62.07 62.08 62.09 62.10 62.11 62.12 62.13 Wibhu Kutanan, Rasmi Shoocongdej, Metawee Srikummool, et al. (2020), "Cultural variation impacts paternal and maternal genetic lineages of the Hmong-Mien and Sino-Tibetan groups from Thailand." European Journal of Human Genetics. https://doi.org/10.1038/s41431-020-0693-x
- ↑ Hartmann et al. 2009, Validation of microarray-based resequencing of 93 worldwide mitochondrial genomes
- ↑ 64.0 64.1 64.2 64.3 64.4 64.5 64.6 64.7 Lippold, S; Xu, H; Ko, A; Li, M; Renaud, G; Butthof, A; Schröder, R; Stoneking, M (2014). "Human paternal and maternal demographic histories: insights from high-resolution Y chromosome and mtDNA sequences". Investig Genet 5: 13. doi:10.1186/2041-2223-5-13. PMID 25254093.
- ↑ 65.0 65.1 65.2 65.3 65.4 Fornarino, SimonaExpression error: Unrecognized word "etal". (2009). "Mitochondrial and Y-chromosome diversity of the Tharus (Nepal): a reservoir of genetic variation". BMC Evolutionary Biology 9: 154. doi:10.1186/1471-2148-9-154. PMID 19573232.
- ↑ 66.0 66.1 Fernando, A. S.; Wanninayaka, A.; Dewage, D.; Karunanayake, E. H.; Rai, N.; Somadeva, R.; Tennekoon, K. H.; Ranasinghe, R. (2023). "The mitochondrial genomes of two Pre-historic Hunter Gatherers in Sri Lanka". Journal of Human Genetics 68 (2): 103–105. doi:10.1038/s10038-022-01099-w. PMID 36450887. https://www.nature.com/articles/s10038-022-01099-w.epdf?sharing_token=lAw-MUL8bjW4ZVcSLKhCUtRgN0jAjWel9jnR3ZoTv0PO2roAz6WGW6t6BlvG2gC_1yxHwCv2QZBaHa6zj062Jw9U2GkSJPVH2hWHoy2P5irDVPl-3o6Tn54B8iHxtRVcSB79WZBTsP8cpck83kb3KoY1sANf-2VCIpHjCDNgBhc%3D.
- ↑ 67.00 67.01 67.02 67.03 67.04 67.05 67.06 67.07 67.08 67.09 67.10 67.11 67.12 67.13 67.14 67.15 67.16 67.17 67.18 Chandrasekar, AExpression error: Unrecognized word "etal". (2009). "Updating Phylogeny of Mitochondrial DNA Macrohaplogroup M in India: Dispersal of Modern Human in South Asian Corridor". PLOS ONE 4 (10): e7447. doi:10.1371/journal.pone.0007447. PMID 19823670. Bibcode: 2009PLoSO...4.7447C.
- ↑ Malyarchuk, B. A.; Perkova, M. A.; Derenko, M. V.; Vanecek, T.; Lazur, J.; Gomolcak, P. (2008). "Mitochondrial DNA Variability in Slovaks, with Application to the Roma Origin". Annals of Human Genetics 72 (2): 228–240. doi:10.1111/j.1469-1809.2007.00410.x. PMID 18205894.
- ↑ Kutanan,W., Kampuansai,J., Brunelli,A., Ghirotto,S., Pittayaporn,P., Ruangchai,S., Schroder,R., Macholdt,E., Srikummool,M., Kangwanpong,D., Hubner,A., Arias,L. and Stoneking,M., "New insights from Thailand into the maternal genetic history of Mainland Southeast Asia." Eur. J. Hum. Genet. (2018).
- ↑ Peng (2011). "Tracing the legacy of the early Hainan Islanders - a perspective from mitochondrial DNA". BMC Evolutionary Biology 11: 46. doi:10.1186/1471-2148-11-46. PMID 21324107.
- ↑ Sukernik, Rem I.; Volodko, Natalia V.; Mazunin, Ilya O.; Eltsov, Nikolai P.; Dryomov, Stanislav V.; Starikovskaya, Elena B. (2012). "Mitochondrial Genome Diversity in the Tubalar, Even, and Ulchi: Contribution to Prehistory of Native Siberians and Their Affinities to Native Americans". American Journal of Physical Anthropology 148 (1): 123–138. doi:10.1002/ajpa.22050. PMID 22487888.
- ↑ 72.0 72.1 Fedorova, Sardana AExpression error: Unrecognized word "etal". (2013). "Autosomal and uniparental portraits of the native populations of Sakha (Yakutia): implications for the peopling of Northeast Eurasia". BMC Evolutionary Biology 13: 127. doi:10.1186/1471-2148-13-127. PMID 23782551.
- ↑ 73.00 73.01 73.02 73.03 73.04 73.05 73.06 73.07 73.08 73.09 73.10 73.11 73.12 73.13 73.14 73.15 73.16 Duggan, ATExpression error: Unrecognized word "etal". (2013). "Investigating the Prehistory of Tungusic Peoples of Siberia and the Amur-Ussuri Region with Complete mtDNA Genome Sequences and Y-chromosomal Markers". PLOS ONE 8 (12): e83570. doi:10.1371/journal.pone.0083570. PMID 24349531. Bibcode: 2013PLoSO...883570D.
- ↑ 74.0 74.1 74.2 74.3 Hongbin Yao, Mengge Wang, Xing Zou, et al., "New insights into the fine-scale history of western-eastern admixture of the northwestern Chinese population in the Hexi Corridor via genome-wide genetic legacy." Mol Genet Genomics 2021 Mar 1. doi: 10.1007/s00438-021-01767-0.
- ↑ 75.0 75.1 Tanaka, MasashiExpression error: Unrecognized word "etal". (October 2004). "Mitochondrial Genome Variation in Eastern Asia and the Peopling of Japan". Genome Res. 14 (10): 1832–1850. doi:10.1101/gr.2286304. PMID 15466285.
- ↑ Chen J, He G, Ren Z, Wang Q, Liu Y, Zhang H, Yang M, Zhang H, Ji J, Zhao J, Guo J, Chen J, Zhu K, Yang X, Wang R, Ma H, Tao L, Liu Y, Shen Q, Yang W, Wang C-C, and Huang J (2022), "Fine-Scale Population Admixture Landscape of Tai–Kadai-Speaking Maonan in Southwest China Inferred From Genome-Wide SNP Data." Front. Genet. 13:815285. doi: 10.3389/fgene.2022.815285
- ↑ Basnet, Rajdip; Rai, Niraj; Tamang, Rakesh; Awasthi, Nagendra Prasad; Pradhan, Isha; Parajuli, Pawan; Kashyap, Deepak; Reddy, Alla Govardhan et al. (2022-10-15). "The matrilineal ancestry of Nepali populations" (in en). Human Genetics 142 (2): 167–180. doi:10.1007/s00439-022-02488-z. ISSN 0340-6717. PMID 36242641. https://link.springer.com/10.1007/s00439-022-02488-z.
- ↑ Zhang, Xiaoming; Ji, Xueping; Li, Chunmei; Yang, Tingyu; Huang, Jiahui; Zhao, Yinhui; Wu, Yun; Ma, Shiwu et al. (July 2022). "A Late Pleistocene human genome from Southwest China". Current Biology 32 (14): 3095–3109.e5. doi:10.1016/j.cub.2022.06.016. ISSN 0960-9822. PMID 35839766.
- ↑ 79.00 79.01 79.02 79.03 79.04 79.05 79.06 79.07 79.08 79.09 79.10 79.11 79.12 79.13 79.14 79.15 79.16 79.17 79.18 79.19 79.20 79.21 79.22 79.23 79.24 79.25 79.26 79.27 79.28 79.29 79.30 79.31 79.32 79.33 79.34 79.35 79.36 Behar et al., 2012b
- ↑ 80.0 80.1 80.2 Derenko, MExpression error: Unrecognized word "etal". (2012). "Complete Mitochondrial DNA Analysis of Eastern Eurasian Haplogroups Rarely Found in Populations of Northern Asia and Eastern Europe". PLOS ONE 7 (2): e32179. doi:10.1371/journal.pone.0032179. PMID 22363811. Bibcode: 2012PLoSO...732179D.
- ↑ Juras, A.; Krzewinska, M.; Nikitin, A.G.; Ehler, E.; Chylenski, M.; Lukasik, S.; Krenz-Niedbala, M.; Sinika, V. et al. (2017). "Diverse origin of mitochondrial lineages in Iron Age Black Sea Scythians". Sci Rep 7: 43950. doi:10.1038/srep43950. PMID 28266657. Bibcode: 2017NatSR...743950J.
- ↑ 82.0 82.1 82.2 Ko, Albert Min-Shan; Chen, Chung-Yu; Fu, Qiaomei; Delfin, Frederick; Li, Mingkun; Chiu, Hung-Lin; Stoneking, Mark; Ko, Ying-Chin (2014). "Early Austronesians: into and out of Taiwan". The American Journal of Human Genetics 94 (3): 426–436. doi:10.1016/j.ajhg.2014.02.003. PMID 24607387.
- ↑ Hwan Young Lee, Ji-Eun Yoo, Myung Jin Park, Ukhee Chung, Chong-Youl Kim, and Kyoung-Jin Shin, "East Asian mtDNA haplogroup determination in Koreans: Haplogroup-level coding region SNP analysis and subhaplogroup-level control region sequence analysis." Electrophoresis (2006). DOI 10.1002/elps.200600151.
- ↑ 84.00 84.01 84.02 84.03 84.04 84.05 84.06 84.07 84.08 84.09 84.10 84.11 84.12 84.13 84.14 84.15 84.16 84.17 84.18 84.19 84.20 84.21 84.22 84.23 84.24 84.25 84.26 84.27 84.28 84.29 84.30 84.31 84.32 MtDNA Haplotree at Family Tree DNA
- ↑ 85.0 85.1 85.2 Dandan Yu, Xiaoyun Jia, A-Mei Zhang, Shiqiang Li, Yang Zou, Qingjiong Zhang, and Yong-Gang Yao (2010), "Mitochondrial DNA Sequence Variation and Haplogroup Distribution in Chinese Patients with LHON and m.14484T.C." PLoS ONE 5(10): e13426. doi:10.1371/journal.pone.0013426
- ↑ Yanli Ji, A-Mei Zhang, Xiaoyun Jia, et al. (2008), "Mitochondrial DNA Haplogroups M7b102 and M8a Affect Clinical Expression of Leber Hereditary Optic Neuropathy in Chinese Families with the m.11778G/A Mutation." The American Journal of Human Genetics 83, 760–768, December 12, 2008. DOI 10.1016/j.ajhg.2008.11.002.
- ↑ 87.0 87.1 87.2 87.3 87.4 87.5 87.6 87.7 Dan Zhao, Yingying Ding, Haijiang Lin, Xiaoxiao Chen, Weiwei Shen, Meiyang Gao, Qian Wei, Sujuan Zhou, Xing Liu, and Na He (2019), "Mitochondrial Haplogroups N9 and G Are Associated with Metabolic Syndrome Among Human Immunodeficiency Virus-Infected Patients in China." AIDS Research and Human Retroviruses, Jun 2019, 536-543. http://doi.org/10.1089/aid.2018.0151
- ↑ Dongsheng Lu, Haiyi Lou, Kai Yuan, et al. (2016), "Ancestral Origins and Genetic History of Tibetan Highlanders." The American Journal of Human Genetics 99, 580–594, September 1, 2016.
- ↑ 89.0 89.1 89.2 89.3 89.4 89.5 89.6 89.7 89.8 Longli Kang, Hong-Xiang Zheng, Menghan Zhang, et al. (2016), "MtDNA analysis reveals enriched pathogenic mutations in Tibetan highlanders." Scientific Reports | 6:31083 | DOI: 10.1038/srep31083.
- ↑ 90.0 90.1 90.2 90.3 Monika Summerer, Jürgen Horst, Gertraud Erhart, et al. (2014), "Large-scale mitochondrial DNA analysis in Southeast Asia reveals evolutionary effects of cultural isolation in the multi-ethnic population of Myanmar." BMC Evolutionary Biology 2014, 14:17. http://www.biomedcentral.com/1471-2148/14/17
- ↑ Max Ingman and Ulf Gyllensten (2007), "Rate variation between mitochondrial domains and adaptive evolution in humans." Human Molecular Genetics, 2007, Vol. 16, No. 19, 2281–2287. doi:10.1093/hmg/ddm180
- ↑ 92.00 92.01 92.02 92.03 92.04 92.05 92.06 92.07 92.08 92.09 92.10 92.11 Pankratov, V., Litvinov, S., Kassian, A., et al., "East Eurasian ancestry in the middle of Europe: genetic footprints of Steppe nomads in the genomes of Belarusian Lipka Tatars." Sci Rep 6, 30197 (2016). https://doi.org/10.1038/srep30197
- ↑ Rebecca S. Just, Melissa K. Scheible, Spence A. Fast, et al. (2015), "Full mtGenome reference data: Development and characterization of 588 forensic-quality haplotypes representing three U.S. populations." Forensic Science International: Genetics 14 (2015) 141–155. http://dx.doi.org/10.1016/j.fsigen.2014.09.021
- ↑ 94.00 94.01 94.02 94.03 94.04 94.05 94.06 94.07 94.08 94.09 94.10 94.11 94.12 94.13 Wibhu Kutanan, Jatupol Kampuansai, Andrea Brunelli, et al. (2018), "New insights from Thailand into the maternal genetic history of Mainland Southeast Asia." European Journal of Human Genetics (2018) 26:898–911. https://doi.org/10.1038/s41431-018-0113-7
- ↑ 95.0 95.1 Qing-Peng Kong, Yong-Gang Yao, Chang Sun, et al. (2003), "Phylogeny of East Asian Mitochondrial DNA Lineages Inferred from Complete Sequences." Am. J. Hum. Genet. 73:671–676, 2003.
- ↑ 96.0 96.1 96.2 Duong,N.T., Macholdt,E., Ton,N.D., et al., "Complete human mtDNA genome sequences from Vietnam and the phylogeography of Mainland Southeast Asia." Sci Rep 8 (1), 11651 (2018).
- ↑ Yang Zou, Xiaoyun Jia, A-Mei Zhang, et al. (2010), "The MT-ND1 and MT-ND5 genes are mutational hotspots for Chinese families with clinical features of LHON but lacking the three primary mutations." Biochemical and Biophysical Research Communications Volume 399, Issue 2, 20 August 2010, Pages 179-185. https://doi.org/10.1016/j.bbrc.2010.07.051
- ↑ 98.0 98.1 98.2 98.3 98.4 98.5 98.6 98.7 Jiang,C., Cui,J., Liu,F., Gao,L., Luo,Y., Li,P., Guan,L. and Gao,Y., "Mitochondrial DNA 10609T Promotes Hypoxia-Induced Increase of Intracellular ROS and Is a Risk Factor of High Altitude Polycythemia." PLoS ONE 9 (1), E87775 (2014).
- ↑ Kong QP, Sun C, Wang HW, Zhao M, Wang WZ, Zhong L, Hao XD, Pan H, Wang SY, Cheng YT, Zhu CL, Wu SF, Liu LN, Jin JQ, Yao YG, Zhang YP. "Large-scale mtDNA screening reveals a surprising matrilineal complexity in east Asia and its implications to the peopling of the region." Mol Biol Evol. 2011 Jan;28(1):513-22. doi: 10.1093/molbev/msq219. Epub 2010 Aug 16. PMID 20713468.
- ↑ 100.0 100.1 100.2 100.3 Jia Liu, Li-Dong Wang, Yan-Bo Sun, et al. (2012), "Deciphering the Signature of Selective Constraints on Cancerous Mitochondrial Genome." Mol. Biol. Evol. 29(4):1255–1261. doi:10.1093/molbev/msr290
- ↑ 101.00 101.01 101.02 101.03 101.04 101.05 101.06 101.07 101.08 101.09 101.10 101.11 101.12 101.13 101.14 101.15 101.16 101.17 101.18 Miroslava Derenko, Galina Denisova, Boris Malyarchuk, Irina Dambueva, and Boris Bazarov, "Mitogenomic diversity and differentiation of the Buryats." Journal of Human Genetics (2018) 63:71–81. https://doi.org/10.1038/s10038-017-0370-2
- ↑ 102.0 102.1 Boris Malyarchuk, Andrey Litvinov, Miroslava Derenko, et al. (2017), "Mitogenomic diversity in Russians and Poles." FSI Genetics Volume 30, p51-56, September 01, 2017. DOI:https://doi.org/10.1016/j.fsigen.2017.06.003
- ↑ Marchi N, Hegay T, Mennecier P, et al. (2017), "Sex-specific genetic diversity is shaped by cultural factors in Inner Asian human populations." American Journal of Physical Anthropology 2017 Apr;162(4):627-640. DOI: 10.1002/ajpa.23151.
- ↑ 104.0 104.1 Hua-Wei Wang, Xiaoyun Jia, Yanli Ji, et al. (2008), "Strikingly different penetrance of LHON in two Chinese families with primary mutation G11778A is independent of mtDNA haplogroup background and secondary mutation G13708A." Mutation Research 643 (2008) 48–53. doi:10.1016/j.mrfmmm.2008.06.004
- ↑ 105.0 105.1 Mielnik-Sikorska M, Daca P, Malyarchuk B, Derenko M, Skonieczna K, et al. (2013), "The History of Slavs Inferred from Complete Mitochondrial Genome Sequences." PLoS ONE 8(1): e54360. doi:10.1371/journal.pone.0054360
- ↑ Gülşah Merve Kılınç, Natalija Kashuba, Reyhan Yaka, et al. (2018), "Investigating Holocene human population history in North Asia using ancient mitogenomes." Scientific Reports (2018) 8:8969 | DOI:10.1038/s41598-018-27325-0
- ↑ 107.0 107.1 Boris Malyarchuk, Miroslava Derenko, Galina Denisova, and Olga Kravtsova (2010), "Mitogenomic Diversity in Tatars from the Volga-Ural Region of Russia." Mol. Biol. Evol. 27(10):2220–2226. doi:10.1093/molbev/msq065
- ↑ Jun-Hun Loo, Jean A Trejaut, Ju-Chen Yen, et al. (2014), "Mitochondrial DNA association study of type 2 diabetes with or without ischemic stroke in Taiwan." BMC Res Notes 2014; 7: 223. Published online 2014 Apr 9. doi: 10.1186/1756-0500-7-223
- ↑ 109.0 109.1 Dryomov SV, Nazhmidenova AM, Starikovskaya EB, Shalaurova SA, Rohland N, Mallick S, et al. (2021), "Mitochondrial genome diversity on the Central Siberian Plateau with particular reference to the prehistory of northernmost Eurasia." PLoS ONE 16(1):e0244228. https://doi.org/10.1371/journal. pone.0244228
- ↑ 110.0 110.1 Wang, C. Y., Li, H., Hao, X. D., Liu, J., Wang, J. X., Wang, W. Z., Kong, Q. P., & Zhang, Y. P. (2011). "Uncovering the profile of somatic mtDNA mutations in Chinese colorectal cancer patients." PloS one, 6(6), e21613. https://doi.org/10.1371/journal.pone.0021613
- ↑ A-Mei Zhang, Xiaoyun Jia, Yong-Gang Yao, and Qingjiong Zhang (2008), "Co-occurrence of A1555G and G11778A in a Chinese family with high penetrance of Leber’s hereditary optic neuropathy." Biochemical and Biophysical Research Communications 376 (2008) 221–224. doi:10.1016/j.bbrc.2008.08.128
- ↑ Choongwon Jeong, Ke Wang, Shevan Wilkin, William Timothy Treal Taylor, Bryan K. Miller, Jan H. Bemmann, Raphaela Stahl, Chelsea Chiovelli, Florian Knolle, Sodnom Ulziibayar, Dorjpurev Khatanbaatar, Diimaajav Erdenebaatar, Ulambayar Erdenebat, Ayudai Ochir, Ganbold Ankhsanaa, Chuluunkhuu Vanchigdash, Battuga Ochir, Chuluunbat Munkhbayar, Dashzeveg Tumen, Alexey Kovalev, Nikolay Kradin, Bilikto A. Bazarov, Denis A. Miyagashev, Prokopiy B. Konovalov, Elena Zhambaltarova, Alicia Ventresca Miller, Wolfgang Haak, Stephan Schiffels, Johannes Krause, Nicole Boivin, Myagmar Erdene, Jessica Hendy, and Christina Warinner, "A Dynamic 6,000-Year Genetic History of Eurasia’s Eastern Steppe." Cell, Volume 183, Issue 4, 2020, Pages 890-904.e29, ISSN 0092-8674, https://doi.org/10.1016/j.cell.2020.10.015.
- ↑ 113.0 113.1 Mian Zhao, Qing-Peng Kong, Hua-Wei Wang, et al. (2009), "Mitochondrial genome evidence reveals successful Late Paleolithic settlement on the Tibetan Plateau." PNAS, vol. 106, no. 50, 21230–21235. http://www.pnas.org/cgi/doi/10.1073/pnas.0907844106
- ↑ Longli Kang, Hong-Xiang Zheng, Feng Chen, et al. (2013), "mtDNA Lineage Expansions in Sherpa Population Suggest Adaptive Evolution in Tibetan Highlands." Molecular Biology and Evolution, Volume 30, Issue 12, December 2013, Pages 2579–2587, https://doi.org/10.1093/molbev/mst147.
- ↑ 115.0 115.1 115.2 115.3 Kong, Qing-PengExpression error: Unrecognized word "etal". (2006). "Updating the East Asian mtDNA phylogeny: a prerequisite for the identification of pathogenic mutations". Human Molecular Genetics 15 (13): 2076–2086. doi:10.1093/hmg/ddl130. PMID 16714301.
- ↑ Xiaoming Zhang, Chunmei Li, Yanan Zhou, et al. (2020), "A Matrilineal Genetic Perspective of Hanging Coffin Custom in Southern China and Northern Thailand." iScience 23, 101032, April 24, 2020. https://doi.org/10.1016/j.isci.2020.101032
- ↑ Dryomov, S.V., Starikovskaya, E.B., Nazhmidenova, A.M. et al. Genetic legacy of cultures indigenous to the Northeast Asian coast in mitochondrial genomes of nearly extinct maritime tribes. BMC Evol Biol 20, 83 (2020). https://doi.org/10.1186/s12862-020-01652-1
- ↑ Cheng-Ye Wang and Zhong-Bao Zhao (2012), "Somatic mtDNA mutations in lung tissues of pesticide-exposed fruit growers." Toxicology 291 (2012) 51– 55. doi:10.1016/j.tox.2011.10.018
- ↑ Ingman, M.; Kaessmann, H.; Paabo, S.; Gyllensten, U. (2000). "Mitochondrial genome variation and the origin of modern humans". Nature 408 (6813): 708–713. doi:10.1038/35047064. PMID 11130070. Bibcode: 2000Natur.408..708I.
- ↑ Yao, Yong-GangExpression error: Unrecognized word "etal". (2004). "Different Matrilineal Contributions to Genetic Structure of Ethnic Groups in the Silk Road Region in China". Mol. Biol. Evol. 21 (12): 2265–2280. doi:10.1093/molbev/msh238. PMID 15317881.
- ↑ Fedorova, Sardana AExpression error: Unrecognized word "etal". (2013). "Autosomal and uniparental portraits of the native populations of Sakha (Yakutia): implications for the peopling of Northeast Eurasia". BMC Evolutionary Biology 13: 127. doi:10.1186/1471-2148-13-127. PMID 23782551.
- ↑ 122.00 122.01 122.02 122.03 122.04 122.05 122.06 122.07 122.08 122.09 122.10 122.11 122.12 122.13 122.14 122.15 Kutanan, WibhuExpression error: Unrecognized word "etal". (2018). "Contrasting maternal and paternal genetic variation of hunter-gatherer groups in Thailand". Scientific Reports 8 (1): 1536. doi:10.1038/s41598-018-20020-0. PMID 29367746. Bibcode: 2018NatSR...8.1536K.
- ↑ 123.0 123.1 123.2 Scholes, ClarissaExpression error: Unrecognized word "etal". (2011). "Genetic Diversity and Evidence for Population Admixture in Batak Negritos from Palawan". American Journal of Physical Anthropology 146 (1): 62–72. doi:10.1002/ajpa.21544. PMID 21796613.
- ↑ "M24'41 MTree". https://yfull.com/mtree/M24'41/.
- ↑ Brook, Kevin Alan (2022). The Maternal Genetic Lineages of Ashkenazic Jews. Academic Studies Press. p. 81. doi:10.2307/j.ctv33mgbcn. ISBN 978-1644699843.
- ↑ 126.0 126.1 126.2 126.3 126.4 126.5 126.6 126.7 126.8 Jiao-Yang Tian; Hua-Wei Wang; Yu-Chun Li; Wen Zhang; Yong-Gang Yao; Jits van Straten; Martin B. Richards; Qing-Peng Kong (11 February 2015). "A Genetic Contribution from the Far East into Ashkenazi Jews via the Ancient Silk Road". Scientific Reports 5: 8377. doi:10.1038/srep08377. PMID 25669617.
- ↑ Brook, Kevin Alan (2022). The Maternal Genetic Lineages of Ashkenazic Jews. Academic Studies Press. p. 80. doi:10.2307/j.ctv33mgbcn. ISBN 978-1644699843.
- ↑ Malyarchuk, B. et al 2008c, Mitochondrial DNA Variability in Slovaks, with Application to the Roma Origin
- ↑ "M39'70 MTree". https://yfull.com/mtree/M39'70/.
- ↑ "M42'74 MTree". https://yfull.com/mtree/M42'74/.
- ↑ Zhang,X., Qi,X., Yang,Z., et al., "Analysis of mitochondrial genome diversity identifies new and ancient maternal lineages in Cambodian aborigines." Nat Commun 4, 2599 (2013).
- ↑ "M55'77 MTree". https://yfull.com/mtree/M55'77/.
- ↑ "M77 MTree". https://yfull.com/mtree/M77/.
- ↑ "M62'68 MTree". https://yfull.com/mtree/M62'68/.
- ↑ "M68 MTree". https://yfull.com/mtree/M68/.
- ↑ 136.0 136.1 Wibhu Kutanan, Jatupol Kampuansai, Piya Changmai, et al. (2018), "Contrasting maternal and paternal genetic variation of hunter-gatherer groups in Thailand." Scientific Reports volume 8, Article number: 1536. doi:10.1038/s41598-018-20020-0
- ↑ "M73'79 MTree". https://yfull.com/mtree/M73'79/.
- ↑ "M73 MTree". https://yfull.com/mtree/M73.
- ↑ "M79 MTree". https://yfull.com/mtree/M79/.
- ↑ Comas et al. (2004), Admixture, migrations, and dispersals in Central Asia: evidence from maternal DNA lineages, European Journal of Human Genetics (2004) 12, 495–504.
External links
- General
- Ian Logan's Mitochondrial DNA Site
- The India DNA geographical project at Family Tree DNA
- The China DNA geographical project at Family Tree DNA
- Haplogroup M
- Mannis van Oven's PhyloTree.org - mtDNA subtree M
- Spread of Haplogroup M, from National Geographic
- Tree of M haplogroup as for 2006
- Haplogroup M (mtDNA) interest group on Facebook
- Another tree emphasizing the Andamanese and Nicobarese populations in comparison with other peoples with high M presence
- K.Tharanghaj et al. In situ origin of deep rooting lineages of mitochondrial Macrohaplogroup M in India (PDF document)
 |

