Biology:Henipavirus
| Henipavirus | |
|---|---|
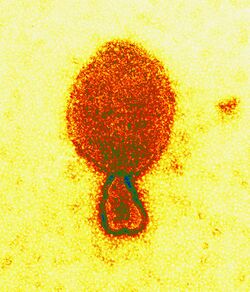
| |
| Colored transmission electron micrograph of a Hendra henipavirus virion (ca. 300 nm length) | |
| Virus classification | |
| Script error: No such module "Taxobox ranks".: | Virus |
| Script error: No such module "Taxobox ranks".: | Riboviria |
| Script error: No such module "Taxobox ranks".: | Orthornavirae |
| Script error: No such module "Taxobox ranks".: | Negarnaviricota |
| Script error: No such module "Taxobox ranks".: | Monjiviricetes |
| Script error: No such module "Taxobox ranks".: | Mononegavirales |
| Script error: No such module "Taxobox ranks".: | Paramyxoviridae |
| Script error: No such module "Taxobox ranks".: | Orthoparamyxovirinae |
| Script error: No such module "Taxobox ranks".: | Henipavirus |
| Species | |
| |
Henipavirus is a genus of negative-strand RNA viruses in the family Paramyxoviridae, order Mononegavirales containing six established species,[1][2] and numerous others still under study.[3] Henipaviruses are naturally harboured by several species of small mammals, notably pteropid fruit bats (flying foxes), microbats of several species,[4] and shrews.[5][6] Henipaviruses are characterised by long genomes and a wide host range. Their recent emergence as zoonotic pathogens capable of causing illness and death in domestic animals and humans is a cause of concern.[7][8]
In 2009, RNA sequences of three novel viruses in phylogenetic relationship to known henipaviruses were detected in African straw-colored fruit bats (Eidolon helvum) in Ghana. The finding of these novel henipaviruses outside Australia and Asia indicates that the region of potential endemicity of henipaviruses may be worldwide.[9] These African henipaviruses are slowly being characterised.[10]
Nipah and Hendra henipaviruses are both considered category C (USDA-HHS overlap) select agents.[11]
Structure
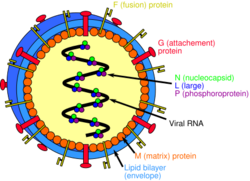
Henipavirions are pleomorphic (variably shaped), ranging in size from 40 to 600 nm in diameter.[12] They possess a lipid membrane overlying a shell of viral matrix protein. At the core is a single helical strand of genomic RNA tightly bound to N (nucleocapsid) protein and associated with the L (large) and P (phosphoprotein) proteins, which provide RNA polymerase activity during replication.
Embedded within the lipid membrane are spikes of F (fusion) protein trimers and G (attachment) protein tetramers. The function of the G protein (except in the case of MojV-G) is to attach the virus to the surface of a host cell via Ephrin B1, B2, or B3, a family of highly conserved mammalian proteins.[13][14][15] The structure of the attachment glycoprotein has been determined by X-ray crystallography.[16] The F protein fuses the viral membrane with the host cell membrane, releasing the virion contents into the cell. It also causes infected cells to fuse with neighbouring cells to form large, multinucleated syncytia.
Genome
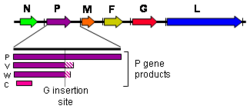
As all mononegaviral genomes, Hendra virus and Nipah virus genomes are non-segmented, single-stranded negative-sense RNA. Both genomes are 18.2 kb in length and contain six genes corresponding to six structural proteins.[17]
In common with other members of the Paramyxoviridae family, the number of nucleotides in the henipavirus genome is a multiple of six, consistent with what is known as the 'rule of six'.[18][19] Deviation from the rule of six, through mutation or incomplete genome synthesis, leads to inefficient viral replication, probably due to structural constraints imposed by the binding between the RNA and the N protein.
Three additional protein products are produced from the henipavirus P gene: V, W, and C. The V and W proteins are generated through an unusual process called RNA editing. This specific process in henipaviruses involves the insertion of extra guanosine residues into the P gene mRNA prior to translation. The addition of a single guanosine results in production of V, and the addition of two guanosines residues produces W.[20] The C protein is not produced through RNA editing but instead by leaky scanning of the host cell ribosome during translation of viral mRNA. P, V, and W possess an alternate open reading frame which results in production of C. P, V, W, and C are known to disrupt the host innate antiviral immune response through several different mechanisms.[21] P, V, and W contain STAT1 binding domains, and act as interferon antagonists by sequestering STAT1 in the nucleus and cytoplasm.[22] The C protein controls the early pro-inflammatory response and is also known to promote the viral budding process via a ESCRT-dependent pathway.[23][24]
Life cycle
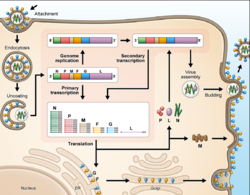
Cell receptor ephrin-B2, which is located on epithelial cells around smaller arteries, neurons, and smooth muscle cells, is targeted by the viral protein G.[25] Once the protein G binds to ephrin-B2, the viral protein F facilitates fusion with the host cell membrane and releases viral RNA into the host cell cytoplasm.[26] Upon entry, transcription of viral mRNA takes place using the viral RNA as a template. This process is started and stopped by the polymerase complex. Viral proteins are gathering in the cell as transcription occurs until the polymerase complex stops transcription and starts genome replication. Transcription of the viral RNA makes positive sense strands of RNA, which are then used as templates to make more negative sense viral RNA . Genome replication is halted before the viral particles can assemble to make a virion. Once the cell membrane is ready, new virions exit the host cell through budding.[27]
Vaccine
Henipaviruses have high mortality rates in mammalian hosts, both human and animal. Because of this, there is a need for immunization against HeV and NiV. The World Health Organization has classified henipaviral agents as R&D Blueprint Priority Pathogens, indicating that they pose a significant risk due to their epidemic potential.[28] The broad species tropism of NiV and HeV have resulted in mortality in livestock species in addition to humans, and as a result veterinary vaccines are in various stages of development or licensure. EquiVac HeV, a veterinary vaccine for horses was licensed in Australia in 2012.[29][30] A number of experimental vaccines designed for humans are in preclinical development, but none have yet been licensed. A soluble HeV attachment glycoprotein vaccine designed to protect against NiV completed a phase I clinical trial in November 2022, but results have not yet been published.[31]
The primary mechanism of protection against NiV and HeV induced by vaccination is thought to be neutralizing antibodies.[32] However, a number of preclinical vaccine studies in animal models of disease have identified that the cell-mediated immune response including CD8+ and CD4+ T-cells may play a role in protection.[33]
Causes of emergence
The emergence of henipaviruses parallels the emergence of other zoonotic viruses in recent decades. SARS coronavirus, Australian bat lyssavirus, Menangle virus, Marburg virus, COVID 19 and possibly Ebola viruses are also harboured by bats, and are capable of infecting a variety of other species. The emergence of each of these viruses has been linked to an increase in contact between bats and humans, sometimes involving an intermediate domestic animal host. The increased contact is driven both by human encroachment into the bats' territory (in the case of Nipah, specifically pigpens in said territory) and by movement of bats towards human populations due to changes in food distribution and loss of habitat.
There is evidence that habitat loss for flying foxes, both in South Asia and Australia (particularly along the east coast) as well as encroachment of human dwellings and agriculture into the remaining habitats, is creating greater overlap of human and flying fox distributions.[34]
Taxonomy
| Genus | Species | Virus (Abbreviation) |
| Henipavirus | Cedar henipavirus | Cedar virus (CedV) |
| Ghanaian bat henipavirus | Kumasi virus (KV) | |
| Hendra henipavirus | Hendra virus (HeV) | |
| Mojiang henipavirus | Mòjiāng virus (MojV)[3] | |
| Nipah henipavirus | Nipah virus (NiV) | |
| Langya henipavirus | Langya virus (LayV)[6][36] |
See also
- Animal viruses
- Paramyxovirus
References
- ↑ Rima, B; Balkema-Buschmann, A; Dundon, WG; Duprex, WP; Easton, A; Fouchier, R; Kurath, G; Lamb, R et al. (December 2019). "ICTV Virus Taxonomy Profile: Paramyxoviridae.". The Journal of General Virology 100 (12): 1593–1594. doi:10.1099/jgv.0.001328. PMID 31609197.
- ↑ "ICTV Report Paramyxoviridae". http://www.ictv.global/report/paramyxoviridae.
- ↑ 3.0 3.1 Wu, Zhiqiang (2014). "Novel Henipa-like Virus, Mojiang Paramyxovirus, in Rats, China, 2012". Emerging Infectious Diseases 20 (6): 1064–1066. doi:10.3201/eid2006.131022. PMID 24865545.
- ↑ Li, Y; Wang, J; Hickey, AC; Zhang, Y; Li, Y; Wu, Y; Zhang, Huajun et al. (December 2008). "Antibodies to Nipah or Nipah-like viruses in bats, China [letter"]. Emerging Infectious Diseases 14 (12): 1974–6. doi:10.3201/eid1412.080359. PMID 19046545.
- ↑ Cheng, Amy (10 August 2022). "New Langya virus that may have spilled over from animals infects dozens". The Washington Post. https://www.washingtonpost.com/health/2022/08/10/langya-virus-china-shrews-henipavirus/.
- ↑ 6.0 6.1 Zhang, Xiao-Ai (2022). "A Zoonotic Henipavirus in Febrile Patients in China". The New England Journal of Medicine 387 (5): 470–472. doi:10.1056/NEJMc2202705. PMID 35921459.
- ↑ Sawatsky (2008). "Hendra and Nipah Virus". Animal Viruses: Molecular Biology. Caister Academic Press. ISBN 978-1-904455-22-6. http://www.horizonpress.com/avir.
- ↑ "Nipah yet to be confirmed, 86 under observation: Shailaja" (in en). https://english.manoramaonline.com/news/kerala/2019/06/03/nipah-case-kochi-thodupuzha-kk-shilaja.html.
- ↑ Markotter, Wanda, ed (2009). "Henipavirus RNA in African Bats". PLOS ONE 4 (7): e6367. doi:10.1371/journal.pone.0006367. PMID 19636378. Bibcode: 2009PLoSO...4.6367D.
- ↑ Drexler JF, Corman VM (2012). "Bats host major mammalian paramyxoviruses". Nat Commun 3: 796. doi:10.1038/ncomms1796. PMID 22531181. Bibcode: 2012NatCo...3..796D.
- ↑ "Federal Select Agent Program" (in en-us). 2021-01-08. https://www.selectagents.gov/index.htm.
- ↑ "Ultrastructure of Hendra virus and Nipah virus within cultured cells and host animals". Microbes and Infection 3 (4): 297–306. 2001. doi:10.1016/S1286-4579(01)01383-1. PMID 11334747.
- ↑ Bonaparte, M; Dimitrov, A; Bossart, K (2005). "Ephrin-B2 ligand is a functional receptor for Hendra virus and Nipah virus". Proceedings of the National Academy of Sciences 102 (30): 10652–7. doi:10.1073/pnas.0504887102. PMID 15998730. Bibcode: 2005PNAS..10210652B.
- ↑ "EphrinB2 is the entry receptor for Nipah virus, an emergent deadly paramyxovirus". Nature 436 (7049): 401–5. 2005. doi:10.1038/nature03838. PMID 16007075. Bibcode: 2005Natur.436..401N.
- ↑ Bowden, Thomas A.; Crispin, Max; Jones, E. Yvonne; Stuart, David I. (1 October 2010). "Shared paramyxoviral glycoprotein architecture is adapted for diverse attachment strategies". Biochemical Society Transactions 38 (5): 1349–1355. doi:10.1042/BST0381349. PMID 20863312.
- ↑ Bowden, Thomas A.; Crispin, Max; Harvey, David J.; Aricescu, A. Radu; Grimes, Jonathan M.; Jones, E. Yvonne; Stuart, David I. (1 December 2008). "Crystal Structure and Carbohydrate Analysis of Nipah Virus Attachment Glycoprotein: a Template for Antiviral and Vaccine Design". Journal of Virology 82 (23): 11628–11636. doi:10.1128/JVI.01344-08. PMID 18815311.
- ↑ "Molecular biology of Hendra and Nipah viruses". Microbes and Infection 3 (4): 279–87. 2001. doi:10.1016/S1286-4579(01)01381-8. PMID 11334745.
- ↑ Halpin, Kim; Bankamp, Bettina; Harcourt, Brian H.; Bellini, William J.; Rota, Paul A. (2004). "Nipah virus conforms to the rule of six in a minigenome replication assay". Journal of General Virology 85 (3): 701–707. doi:10.1099/vir.0.19685-0. ISSN 1465-2099. PMID 14993656.
- ↑ Kolakofsky, D; Pelet, T; Garcin, D; Hausmann, S; Curran, J; Roux, L (February 1998). "Paramyxovirus RNA synthesis and the requirement for hexamer genome length: the rule of six revisited". Journal of Virology 72 (2): 891–9. doi:10.1128/JVI.72.2.891-899.1998. PMID 9444980.
- ↑ Shaw, Megan L. (December 2009). "Henipaviruses Employ a Multifaceted Approach to Evade the Antiviral Interferon Response" (in en). Viruses 1 (3): 1190–1203. doi:10.3390/v1031190. ISSN 1999-4915. PMID 21994589.
- ↑ Lawrence, Philip; Escudero-Pérez, Beatriz (2022-04-29). "Henipavirus Immune Evasion and Pathogenesis Mechanisms: Lessons Learnt from Natural Infection and Animal Models". Viruses 14 (5): 936. doi:10.3390/v14050936. ISSN 1999-4915. PMID 35632678.
- ↑ Shaw, Megan L.; García-Sastre, Adolfo; Palese, Peter; Basler, Christopher F. (June 2004). "Nipah virus V and W proteins have a common STAT1-binding domain yet inhibit STAT1 activation from the cytoplasmic and nuclear compartments, respectively". Journal of Virology 78 (11): 5633–5641. doi:10.1128/JVI.78.11.5633-5641.2004. ISSN 0022-538X. PMID 15140960.
- ↑ Lo, Michael K.; Peeples, Mark E.; Bellini, William J.; Nichol, Stuart T.; Rota, Paul A.; Spiropoulou, Christina F. (2012-10-19). "Distinct and Overlapping Roles of Nipah Virus P Gene Products in Modulating the Human Endothelial Cell Antiviral Response" (in en). PLOS ONE 7 (10): e47790. doi:10.1371/journal.pone.0047790. ISSN 1932-6203. PMID 23094089. Bibcode: 2012PLoSO...747790L.
- ↑ Park, Arnold; Yun, Tatyana; Vigant, Frederic; Pernet, Olivier; Won, Sohui T.; Dawes, Brian E.; Bartkowski, Wojciech; Freiberg, Alexander N. et al. (2016-05-20). "Nipah Virus C Protein Recruits Tsg101 to Promote the Efficient Release of Virus in an ESCRT-Dependent Pathway" (in en). PLOS Pathogens 12 (5): e1005659. doi:10.1371/journal.ppat.1005659. ISSN 1553-7374. PMID 27203423.
- ↑ Bonaparte, Matthew I.; Dimitrov, Antony S.; Bossart, Katharine N.; Crameri, Gary; Mungall, Bruce A.; Bishop, Kimberly A.; Choudhry, Vidita; Dimitrov, Dimiter S. et al. (2005-07-05). "Ephrin-B2 ligand is a functional receptor for Hendra virus and Nipah virus". Proceedings of the National Academy of Sciences 102 (30): 10652–10657. doi:10.1073/pnas.0504887102. ISSN 0027-8424. PMID 15998730. Bibcode: 2005PNAS..10210652B.
- ↑ Zuckerman, Arie J. (1996-06-10). "Fields virology, 3rd edn. (two vol. set): Edited by B.N. Fields, D.M. Knipe, P.M. Howley, R.M. Chanock, J.L. Melnick, T.P. Monath, B. Roizman and S.E. Straus, Lippincott-Raven, Philadelphia, PA. 1996. 3216 pp. $339.50 (hc). ISBN 0 7817 0253 4" (in en). FEBS Letters 388 (1): 88. doi:10.1016/0014-5793(96)88179-8.
- ↑ Rota, Paul A.; Lo, Michael K. (2012), Lee, Benhur; Rota, Paul A., eds., "Molecular Virology of the Henipaviruses", Henipavirus (Berlin, Heidelberg: Springer Berlin Heidelberg) 359: pp. 41–58, doi:10.1007/82_2012_211, ISBN 978-3-642-29818-9, PMID 22552699
- ↑ "Prioritizing diseases for research and development in emergency contexts" (in en). https://www.who.int/activities/prioritizing-diseases-for-research-and-development-in-emergency-contexts.
- ↑ "Equivac® HeV" (in en). https://www.zoetis.com.au/all-products/portal-site/equivac-hev.aspx.
- ↑ Halpin, Kim; Graham, Kerryne; Durr, Peter A. (2021-07-02). "Sero-Monitoring of Horses Demonstrates the Equivac® HeV Hendra Virus Vaccine to Be Highly Effective in Inducing Neutralising Antibody Titres". Vaccines 9 (7): 731. doi:10.3390/vaccines9070731. ISSN 2076-393X. PMID 34358146.
- ↑ Auro Vaccines LLC (2022-11-16). A Phase 1 Randomized, Placebo-controlled, Observer-blind Trial to Assess the Safety and Immunogenicity of a Nipah Vaccine, HeV-sG-V (Hendra Virus Soluble Glycoprotein Vaccine), in Healthy Adults. PATH, Coalition for Epidemic Preparedness Innovations, Cincinnati Children's Hospital Medical Center (CCHMC). https://clinicaltrials.gov/ct2/show/NCT04199169.
- ↑ Amaya, Moushimi; Broder, Christopher C. (2020-09-29). "Vaccines to Emerging Viruses: Nipah and Hendra" (in en). Annual Review of Virology 7 (1): 447–473. doi:10.1146/annurev-virology-021920-113833. ISSN 2327-056X. PMID 32991264.
- ↑ Liew, Yvonne Jing Mei; Ibrahim, Puteri Ainaa S.; Ong, Hui Ming; Chong, Chee Ning; Tan, Chong Tin; Schee, Jie Ping; Gómez Román, Raúl; Cherian, Neil George et al. (June 2022). "The Immunobiology of Nipah Virus" (in en). Microorganisms 10 (6): 1162. doi:10.3390/microorganisms10061162. ISSN 2076-2607. PMID 35744680.
- ↑ Breed, Andrew C.; Field, Hume E.; Epstein, Jonathan H.; Daszak, Peter (August 2006). "Emerging henipaviruses and flying foxes - Conservation and management perspectives". Biological Conservation 131 (2): 211–220. doi:10.1016/j.biocon.2006.04.007. ISSN 0006-3207. PMID 32226079.
- ↑ Amarasinghe, Gaya K.; Bào, Yīmíng; Basler, Christopher F.; Bavari, Sina; Beer, Martin; Bejerman, Nicolás; Blasdell, Kim R.; Bochnowski, Alisa et al. (7 April 2017). "Taxonomy of the order Mononegavirales: update 2017". Archives of Virology 162 (8): 2493–2504. doi:10.1007/s00705-017-3311-7. ISSN 1432-8798. PMID 28389807.
- ↑ "Zoonotic Langya virus found in China, CDC says - Taipei Times". 9 August 2022. https://www.taipeitimes.com/News/taiwan/archives/2022/08/09/2003783223.
External links
- ICTV Report: Paramyxoviridae
- Disease card
- ViralZone: Henipavirus
- Henipavirus – Henipavirus Ecology Research Group (HERG) INFO
- Virus Pathogen Database and Analysis Resource (ViPR): Paramyxoviridae
Wikidata ☰ Q1481398 entry
 |
