Biology:Ruminant
| Ruminants | |
|---|---|

| |
| Scientific classification | |
| Kingdom: | Animalia |
| Phylum: | Chordata |
| Class: | Mammalia |
| Order: | Artiodactyla |
| Clade: | Cetruminantia |
| Clade: | Ruminantiamorpha Spaulding et al., 2009 |
| Suborder: | Ruminantia Scopoli, 1777 |
| Infraorders | |
Ruminants are herbivorous grazing or browsing artiodactyls belonging to the suborder Ruminantia that are able to acquire nutrients from plant-based food by fermenting it in a specialized stomach prior to digestion, principally through microbial actions. The process, which takes place in the front part of the digestive system and therefore is called foregut fermentation, typically requires the fermented ingesta (known as cud) to be regurgitated and chewed again. The process of rechewing the cud to further break down plant matter and stimulate digestion is called rumination.[2][3] The word "ruminant" comes from the Latin ruminare, which means "to chew over again".
The roughly 200 species of ruminants include both domestic and wild species.[4] Ruminating mammals include cattle, all domesticated and wild bovines, goats, sheep, giraffes, deer, gazelles, and antelopes.[5] It has also been suggested that notoungulates also relied on rumination, as opposed to other atlantogenates that rely on the more typical hindgut fermentation, though this is not entirely certain.[6]
Taxonomically, the suborder Ruminantia is a lineage of herbivorous artiodactyls that includes the most advanced and widespread of the world's ungulates.[7] The suborder Ruminantia includes six different families: Tragulidae, Giraffidae, Antilocapridae, Cervidae, Moschidae, and Bovidae.[4]
Taxonomy and evolution
File:Gazelle rumination - zoom.webm Hofmann and Stewart divided ruminants into three major categories based on their feed type and feeding habits: concentrate selectors, intermediate types, and grass/roughage eaters, with the assumption that feeding habits in ruminants cause morphological differences in their digestive systems, including salivary glands, rumen size, and rumen papillae.[8][9] However, Woodall found that there is little correlation between the fiber content of a ruminant's diet and morphological characteristics, meaning that the categorical divisions of ruminants by Hofmann and Stewart warrant further research.[10]
Also, some mammals are pseudoruminants, which have a three-compartment stomach instead of four like ruminants. The Hippopotamidae (comprising hippopotamuses) are well-known examples. Pseudoruminants, like traditional ruminants, are foregut fermentors and most ruminate or chew cud. However, their anatomy and method of digestion differs significantly from that of a four-chambered ruminant.[5]
Monogastric herbivores, such as rhinoceroses, horses, guinea pigs, and rabbits, are not ruminants, as they have a simple single-chambered stomach. These hindgut fermenters digest cellulose in an enlarged cecum. In smaller hindgut fermenters of the order Lagomorpha (rabbits, hares, and pikas), and Caviomorph rodents (Guinea pigs, capybaras, etc), cecotropes formed in the cecum are passed through the large intestine and subsequently reingested to allow another opportunity to absorb nutrients.
Phylogeny
Ruminantia is a crown group of ruminants within the order Artiodactyla, cladistically defined by Spaulding et al. as "the least inclusive clade that includes Bos taurus (cow) and Tragulus napu (mouse deer)". Ruminantiamorpha is a higher-level clade of artiodactyls, cladistically defined by Spaulding et al. as "Ruminantia plus all extinct taxa more closely related to extant members of Ruminantia than to any other living species."[11] This is a stem-based definition for Ruminantiamorpha, and is more inclusive than the crown group Ruminantia. As a crown group, Ruminantia only includes the last common ancestor of all extant (living) ruminants and their descendants (living or extinct), whereas Ruminantiamorpha, as a stem group, also includes more basal extinct ruminant ancestors that are more closely related to living ruminants than to other members of Artiodactyla. When considering only living taxa (neontology), this makes Ruminantiamorpha and Ruminantia synonymous, and only Ruminantia is used. Thus, Ruminantiamorpha is only used in the context of paleontology. Accordingly, Spaulding grouped some genera of the extinct family Anthracotheriidae within Ruminantiamorpha (but not in Ruminantia), but placed others within Ruminantiamorpha's sister clade, Cetancodontamorpha.[11]
Ruminantia's placement within Artiodactyla can be represented in the following cladogram:[12][13][14][15][16]
| Artiodactyla |
| ||||||||||||||||||||||||||||||
Within Ruminantia, the Tragulidae (mouse deer) are considered the most basal family,[17] with the remaining ruminants classified as belonging to the infraorder Pecora. Until the beginning of the 21st century it was understood that the family Moschidae (musk deer) was sister to Cervidae. However, a 2003 phylogenetic study by Alexandre Hassanin (of National Museum of Natural History, France) and colleagues, based on mitochondrial and nuclear analyses, revealed that Moschidae and Bovidae form a clade sister to Cervidae. According to the study, Cervidae diverged from the Bovidae-Moschidae clade 27 to 28 million years ago.[18] The following cladogram is based on a large-scale genome ruminant genome sequence study from 2019:[19]
| Ruminantia |
| ||||||||||||||||||||||||||||||
Classification
- ORDER ARTIODACTYLA
- Suborder Tylopoda: camels and llamas, 7 living species in 3 genera
- Suborder Suina: pigs and peccaries
- Suborder Cetruminantia: ruminants, whales and hippos
- unranked Ruminantia
- Infraorder Tragulina (paraphyletic)[1]
- Family †Leptomerycidae
- Family †Hypertragulidae
- Family †Praetragulidae
- Family †Gelocidae
- Family †Bachitheriidae
- Family Tragulidae: chevrotains, 6 living species in 4 genera
- Family †Archaeomerycidae
- Family †Lophiomerycidae
- Infraorder Pecora
- Family Cervidae: deer and moose, 49 living species in 16 genera
- Family †Palaeomerycidae
- Family †Dromomerycidae
- Family †Hoplitomerycidae
- Family †Climacoceratidae
- Family Giraffidae: giraffe and okapi, 2 living species in 2 genera
- Family Antilocapridae: pronghorn, one living species in one genus
- Family Moschidae: musk deer, 4 living species in one genus
- Family Bovidae: cattle, goats, sheep, and antelope, 143 living species in 53 genera
- Infraorder Tragulina (paraphyletic)[1]
- unranked Ruminantia
Digestive system of ruminants

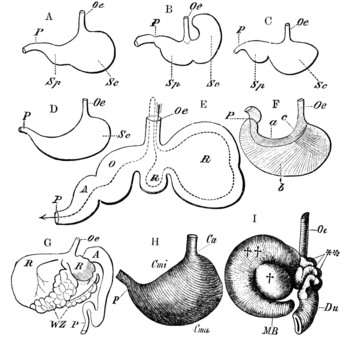
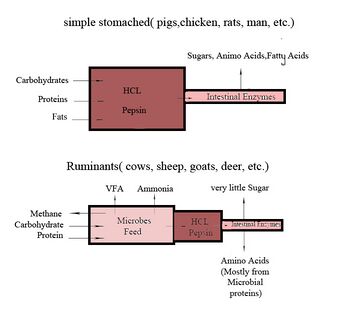
The primary difference between ruminants and nonruminants is that ruminants' stomachs have four compartments:
- rumen—primary site of microbial fermentation
- reticulum
- omasum—receives chewed cud, and absorbs volatile fatty acids
- abomasum—true stomach
The first two chambers are the rumen and the reticulum. These two compartments make up the fermentation vat and are the major site of microbial activity. Fermentation is crucial to digestion because it breaks down complex carbohydrates, such as cellulose, and enables the animal to use them. Microbes function best in a warm, moist, anaerobic environment with a temperature range of 37.7 to 42.2 °C (100 to 108 °F) and a pH between 6.0 and 6.4. Without the help of microbes, ruminants would not be able to use nutrients from forages.[21] The food is mixed with saliva and separates into layers of solid and liquid material.[22] Solids clump together to form the cud or bolus.
The cud is then regurgitated and chewed to completely mix it with saliva and to break down the particle size. Smaller particle size allows for increased nutrient absorption. Fiber, especially cellulose and hemicellulose, is primarily broken down in these chambers by microbes (mostly bacteria, as well as some protozoa, fungi, and yeast) into the three volatile fatty acids (VFAs): acetic acid, propionic acid, and butyric acid. Protein and nonstructural carbohydrate (pectin, sugars, and starches) are also fermented. Saliva is very important because it provides liquid for the microbial population, recirculates nitrogen and minerals, and acts as a buffer for the rumen pH.[21] The type of feed the animal consumes affects the amount of saliva that is produced.
Though the rumen and reticulum have different names, they have very similar tissue layers and textures, making it difficult to visually separate them. They also perform similar tasks. Together, these chambers are called the reticulorumen. The degraded digesta, which is now in the lower liquid part of the reticulorumen, then passes into the next chamber, the omasum. This chamber controls what is able to pass into the abomasum. It keeps the particle size as small as possible in order to pass into the abomasum. The omasum also absorbs volatile fatty acids and ammonia.[21]
After this, the digesta is moved to the true stomach, the abomasum. This is the gastric compartment of the ruminant stomach. The abomasum is the direct equivalent of the monogastric stomach, and digesta is digested here in much the same way. This compartment releases acids and enzymes that further digest the material passing through. This is also where the ruminant digests the microbes produced in the rumen.[21] Digesta is finally moved into the small intestine, where the digestion and absorption of nutrients occurs. The small intestine is the main site of nutrient absorption. The surface area of the digesta is greatly increased here because of the villi that are in the small intestine. This increased surface area allows for greater nutrient absorption. Microbes produced in the reticulorumen are also digested in the small intestine. After the small intestine is the large intestine. The major roles here are breaking down mainly fiber by fermentation with microbes, absorption of water (ions and minerals) and other fermented products, and also expelling waste.[23] Fermentation continues in the large intestine in the same way as in the reticulorumen.
Only small amounts of glucose are absorbed from dietary carbohydrates. Most dietary carbohydrates are fermented into VFAs in the rumen. The glucose needed as energy for the brain and for lactose and milk fat in milk production, as well as other uses, comes from nonsugar sources, such as the VFA propionate, glycerol, lactate, and protein. The VFA propionate is used for around 70% of the glucose and glycogen produced and protein for another 20% (50% under starvation conditions).[24][25]
Abundance, distribution, and domestication
Wild ruminants number at least 75 million[26] and are native to all continents except Antarctica and Australia.[4] Nearly 90% of all species are found in Eurasia and Africa.[26] Species inhabit a wide range of climates (from tropic to arctic) and habitats (from open plains to forests).[26]
The population of domestic ruminants is greater than 3.5 billion, with cattle, sheep, and goats accounting for about 95% of the total population. Goats were domesticated in the Near East circa 8000 BC. Most other species were domesticated by 2500 BC., either in the Near East or southern Asia.[26]
Ruminant physiology
Ruminating animals have various physiological features that enable them to survive in nature. One feature of ruminants is their continuously growing teeth. During grazing, the silica content in forage causes abrasion of the teeth. This is compensated for by continuous tooth growth throughout the ruminant's life, as opposed to humans or other nonruminants, whose teeth stop growing after a particular age. Most ruminants do not have upper incisors; instead, they have a thick dental pad to thoroughly chew plant-based food.[27] Another feature of ruminants is the large ruminal storage capacity that gives them the ability to consume feed rapidly and complete the chewing process later. This is known as rumination, which consists of the regurgitation of feed, rechewing, resalivation, and reswallowing. Rumination reduces particle size, which enhances microbial function and allows the digesta to pass more easily through the digestive tract.[21]
Rumen microbiology
Vertebrates lack the ability to hydrolyse the beta [1–4] glycosidic bond of plant cellulose due to the lack of the enzyme cellulase. Thus, ruminants completely depend on the microbial flora, present in the rumen or hindgut, to digest cellulose. Digestion of food in the rumen is primarily carried out by the rumen microflora, which contains dense populations of several species of bacteria, protozoa, sometimes yeasts and other fungi – 1 ml of rumen is estimated to contain 10–50 billion bacteria and 1 million protozoa, as well as several yeasts and fungi.[28]
Since the environment inside a rumen is anaerobic, most of these microbial species are obligate or facultative anaerobes that can decompose complex plant material, such as cellulose, hemicellulose, starch, and proteins. The hydrolysis of cellulose results in sugars, which are further fermented to acetate, lactate, propionate, butyrate, carbon dioxide, and methane.
As bacteria conduct fermentation in the rumen, they consume about 10% of the carbon, 60% of the phosphorus, and 80% of the nitrogen that the ruminant ingests.[29] To reclaim these nutrients, the ruminant then digests the bacteria in the abomasum. The enzyme lysozyme has adapted to facilitate digestion of bacteria in the ruminant abomasum.[30] Pancreatic ribonuclease also degrades bacterial RNA in the ruminant small intestine as a source of nitrogen.[31]
During grazing, ruminants produce large amounts of saliva – estimates range from 100 to 150 litres of saliva per day for a cow.[32] The role of saliva is to provide ample fluid for rumen fermentation and to act as a buffering agent.[33] Rumen fermentation produces large amounts of organic acids, thus maintaining the appropriate pH of rumen fluids is a critical factor in rumen fermentation. After digesta passes through the rumen, the omasum absorbs excess fluid so that digestive enzymes and acid in the abomasum are not diluted.[1]
Tannin toxicity in ruminant animals
Tannins are phenolic compounds that are commonly found in plants. Found in the leaf, bud, seed, root, and stem tissues, tannins are widely distributed in many different species of plants. Tannins are separated into two classes: hydrolysable tannins and condensed tannins. Depending on their concentration and nature, either class can have adverse or beneficial effects. Tannins can be beneficial, having been shown to increase milk production, wool growth, ovulation rate, and lambing percentage, as well as reducing bloat risk and reducing internal parasite burdens.[34]
Tannins can be toxic to ruminants, in that they precipitate proteins, making them unavailable for digestion, and they inhibit the absorption of nutrients by reducing the populations of proteolytic rumen bacteria.[34][35] Very high levels of tannin intake can produce toxicity that can even cause death.[36] Animals that normally consume tannin-rich plants can develop defensive mechanisms against tannins, such as the strategic deployment of lipids and extracellular polysaccharides that have a high affinity to binding to tannins.[34] Some ruminants (goats, deer, elk, moose) are able to consume food high in tannins (leaves, twigs, bark) due to the presence in their saliva of tannin-binding proteins.[37]
Religious importance
The Law of Moses in the Bible allowed the eating of some mammals that had cloven hooves (i.e. members of the order Artiodactyla) and "that chew the cud",[38] a stipulation preserved to this day in Jewish dietary laws.
Other uses
The verb 'to ruminate' has been extended metaphorically to mean to ponder thoughtfully or to meditate on some topic. Similarly, ideas may be 'chewed on' or 'digested'. 'Chew the (one's) cud' is to reflect or meditate. In psychology, "rumination" refers to a pattern of thinking, and is unrelated to digestive physiology.
Ruminants and climate change
Methane is produced by a type of archaea, called methanogens, as described above within the rumen, and this methane is released to the atmosphere. The rumen is the major site of methane production in ruminants.[39] Methane is a strong greenhouse gas with a global warming potential of 86 compared to CO2 over a 20-year period.[40][41][42]
As a by-product of consuming cellulose, cattle belch out methane, there-by returning that carbon sequestered by plants back into the atmosphere. After about 10 to 12 years, that methane is broken down and converted back to CO
2. Once converted to CO
2, plants can again perform photosynthesis and fix that carbon back into cellulose. From here, cattle can eat the plants and the cycle begins once again. In essence, the methane belched from cattle is not adding new carbon to the atmosphere. Rather it is part of the natural cycling of carbon through the biogenic carbon cycle.[43]
In 2010, enteric fermentation accounted for 43% of the total greenhouse gas emissions from all agricultural activity in the world,[44] 26% of the total greenhouse gas emissions from agricultural activity in the U.S., and 22% of the total U.S. methane emissions.[45] The meat from domestically raised ruminants has a higher carbon equivalent footprint than other meats or vegetarian sources of protein based on a global meta-analysis of lifecycle assessment studies.[46] Methane production by meat animals, principally ruminants, is estimated 15–20% global production of methane, unless the animals were hunted in the wild.[47][48] The current U.S. domestic beef and dairy cattle population is around 90 million head, approximately 50% higher than the peak wild population of American bison of 60 million head in the 1700s,[49] which primarily roamed the part of North America that now makes up the United States.
See also
References
- ↑ 1.0 1.1 1.2 Clauss, M.; Rossner, G. E. (2014). "Old world ruminant morphophysiology, life history, and fossil record: exploring key innovations of a diversification sequence". Annales Zoologici Fennici 51 (1–2): 80–94. doi:10.5735/086.051.0210. http://www.zora.uzh.ch/id/eprint/94203/1/AnnZoolFenn_omasum_2014.pdf.
- ↑ "Rumination: The process of foregut fermentation". http://www.ultimateungulate.com/cetartiodactyla/Rumination.html.
- ↑ "Ruminant Digestive System". http://faculty.fortlewis.edu/LASHELL_B/Nutr2-Rumdigestion.pdf.
- ↑ 4.0 4.1 4.2 Fernández, Manuel Hernández; Vrba, Elisabeth S. (2005-05-01). "A complete estimate of the phylogenetic relationships in Ruminantia: a dated species-level supertree of the extant ruminants" (in en). Biological Reviews 80 (2): 269–302. doi:10.1017/s1464793104006670. ISSN 1469-185X. PMID 15921052.
- ↑ 5.0 5.1 Fowler, M.E. (2010). "Medicine and Surgery of Camelids", Ames, Iowa: Wiley-Blackwell. Chapter 1 General Biology and Evolution addresses the fact that camelids (including camels and llamas) are not ruminants, pseudo-ruminants, or modified ruminants.
- ↑ Richard F. Kay, M. Susana Bargo, Early Miocene Paleobiology in Patagonia: High-Latitude Paleocommunities of the Santa Cruz Formation, Cambridge University Press, 11/10/2012
- ↑ "Suborder Ruminatia, the Ultimate Ungulate". http://www.ultimateungulate.com/cetartiodactyla/Ruminantia.html.
- ↑ Ditchkoff, S. S. (2000). "A decade since "diversification of ruminants": has our knowledge improved?". Oecologia 125 (1): 82–84. doi:10.1007/PL00008894. PMID 28308225. Bibcode: 2000Oecol.125...82D. https://fp.auburn.edu/sfws/ditchkoff/PDF%20publications/2000%20-%20Oecologia.pdf.
- ↑ Reinhold R Hofmann, 1989."Evolutionary steps of ecophysiological and diversification of ruminants: a comparative view of their digestive system". Oecologia, 78:443–457
- ↑ Woodall, P. F. (1992-06-01). "An evaluation of a rapid method for estimating digestibility" (in en). African Journal of Ecology 30 (2): 181–185. doi:10.1111/j.1365-2028.1992.tb00492.x. ISSN 1365-2028.
- ↑ 11.0 11.1 Spaulding, M; O'Leary, MA; Gatesy, J (2009). "Relationships of Cetacea (Artiodactyla) among mammals: increased taxon sampling alters interpretations of key fossils and character evolution". PLOS ONE 4 (9): e7062. doi:10.1371/journal.pone.0007062. PMID 19774069. Bibcode: 2009PLoSO...4.7062S.
- ↑ Beck, N.R. (2006). "A higher-level MRP supertree of placental mammals". BMC Evol Biol 6: 93. doi:10.1186/1471-2148-6-93. PMID 17101039.
- ↑ O'Leary, M.A.; Bloch, J.I.; Flynn, J.J.; Gaudin, T.J.; Giallombardo, A.; Giannini, N.P.; Goldberg, S.L.; Kraatz, B.P. et al. (2013). "The Placental Mammal Ancestor and the Post-K-Pg Radiation of Placentals". Science 339 (6120): 662–667. doi:10.1126/science.1229237. PMID 23393258. Bibcode: 2013Sci...339..662O.
- ↑ Song, S.; Liu, L.; Edwards, S.V.; Wu, S. (2012). "Resolving conflict in eutherian mammal phylogeny using phylogenomics and the multispecies coalescent model". Proceedings of the National Academy of Sciences 109 (37): 14942–14947. doi:10.1073/pnas.1211733109. PMID 22930817. Bibcode: 2012PNAS..10914942S.
- ↑ dos Reis, M.; Inoue, J.; Hasegawa, M.; Asher, R.J.; Donoghue, P.C.J.; Yang, Z. (2012). "Phylogenomic datasets provide both precision and accuracy in estimating the timescale of placental mammal phylogeny". Proceedings of the Royal Society B: Biological Sciences 279 (1742): 3491–3500. doi:10.1098/rspb.2012.0683. PMID 22628470.
- ↑ Upham, N.S.; Esselstyn, J.A.; Jetz, W. (2019). "Inferring the mammal tree: Species-level sets of phylogenies for questions in ecology, evolution, and conservation". PLOS Biology 17 (12): e3000494. doi:10.1371/journal.pbio.3000494. PMID 31800571.(see e.g. Fig S10)
- ↑ Kulemzina, Anastasia I.; Yang, Fengtang; Trifonov, Vladimir A.; Ryder, Oliver A.; Ferguson-Smith, Malcolm A.; Graphodatsky, Alexander S. (2011). "Chromosome painting in Tragulidae facilitates the reconstruction of Ruminantia ancestral karyotype". Chromosome Research 19 (4): 531–539. doi:10.1007/s10577-011-9201-z. ISSN 0967-3849. PMID 21445689.
- ↑ Hassanin, A.; Douzery, E. J. P. (2003). "Molecular and morphological phylogenies of Ruminantia and the alternative position of the Moschidae". Systematic Biology 52 (2): 206–28. doi:10.1080/10635150390192726. PMID 12746147. https://www.researchgate.net/publication/10760976.
- ↑ Chen, L.; Qiu, Q.; Jiang, Y.; Wang, K. (2019). "Large-scale ruminant genome sequencing provides insights into their evolution and distinct traits". Science 364 (6446): eaav6202. doi:10.1126/science.aav6202. PMID 31221828. Bibcode: 2019Sci...364.6202C.
- ↑ Russell, J. B. 2002. Rumen Microbiology and its role In Ruminant Nutrition.
- ↑ 21.0 21.1 21.2 21.3 21.4 Rickard, Tony (2002). Dairy Grazing Manual. MU Extension, University of Missouri-Columbia. pp. 7–8.
- ↑ "How do ruminants digest?". The Open University. http://www.open.edu/openlearn/science-maths-technology/science/biology/how-do-ruminants-digest.
- ↑ Meyer. Class Lecture. Animal Nutrition. University of Missouri-Columbia, MO. 16 September 2016
- ↑ William O. Reece (2005). Functional Anatomy and Physiology of Domestic Animals, pages 357–358 ISBN 978-0-7817-4333-4
- ↑ Colorado State University, Hypertexts for Biomedical Science: Nutrient Absorption and Utilization in Ruminants
- ↑ 26.0 26.1 26.2 26.3 Hackmann. T. J., and Spain, J. N. 2010."Ruminant ecology and evolution: Perspectives useful to livestock research and production". Journal of Dairy Science, 93:1320–1334
- ↑ "Dental Anatomy of Ruminants". http://www.vivo.colostate.edu/hbooks/pathphys/digestion/pregastric/cowpage.html.
- ↑ "Fermentation Microbiology and Ecology". http://arbl.cvmbs.colostate.edu/hbooks/pathphys/digestion/herbivores/microbes.html.
- ↑ Callewaert, L.; Michiels, C. W. (2010). "Lysozymes in the animal kingdom". Journal of Biosciences 35 (1): 127–160. doi:10.1007/S12038-010-0015-5. PMID 20413917.
- ↑ Irwin, D. M.; Prager, E. M.; Wilson, A. C. (1992). "Evolutionary genetics of ruminant lysozymes". Animal Genetics 23 (3): 193–202. doi:10.1111/j.1365-2052.1992.tb00131.x. PMID 1503255.
- ↑ Jermann, T. M.; Opitz, J. G.; Stackhouse, J.; Benner, S. A. (1995). "Reconstructing the evolutionary history of the artiodactyl ribonuclease superfamily". Nature 374 (6517): 57–59. doi:10.1038/374057a0. PMID 7532788. Bibcode: 1995Natur.374...57J. http://64.238.189.139/pubs/Reconstructing%20the%20evolutionary%20history%20of%20the%20artiodactyl%20ribonuclease%20superfamily.pdf.
- ↑ Reid, J.T.; Huffman, C.F. (1949). "Some physical and chemical properties of Bovine saliva which may affect rumen digestion and synthesis". Journal of Dairy Science 32 (2): 123–132. doi:10.3168/jds.s0022-0302(49)92019-6. http://www.journalofdairyscience.org/article/S0022-0302%2849%2992019-6/abstract.

- ↑ "Rumen Physiology and Rumination". http://arbl.cvmbs.colostate.edu:80/hbooks/pathphys/digestion/herbivores/rumination.html.
- ↑ 34.0 34.1 34.2 B.R Min, et al (2003) The effect of condensed tannins on the nutrition and health of ruminants fed fresh temperate forages: a review Animal Feed Science and Technology 106(1):3–19
- ↑ Bate-Smith and Swain (1962). "Flavonoid compounds". in Florkin M., Mason H.S.. Comparative biochemistry. III. New York: Academic Press. pp. 75–809.
- ↑ "'Tannins: fascinating but sometimes dangerous molecules' [Cornell University Department of Animal Science? (c) 2018"]. http://www.ansci.cornell.edu/plants/toxicagents/tannin.html.
- ↑ Austin, PJ (1989). "Tannin-binding proteins in saliva of deer and their absence in saliva of sheep and cattle". J Chem Ecol 15 (4): 1335–47. doi:10.1007/BF01014834. PMID 24272016.
- ↑ Leviticus 11:3
- ↑ Asanuma, Narito; Iwamoto, Miwa; Hino, Tsuneo (1999). "Effect of the Addition of Fumarate on Methane Production by Ruminal Microorganisms in Vitro". Journal of Dairy Science 82 (4): 780–787. doi:10.3168/jds.S0022-0302(99)75296-3. PMID 10212465.
- ↑ IPCC Fifth Assessment Report, Table 8.7, Chap. 8, pp. 8–58 (PDF)
- ↑ Shindell, D. T.; Faluvegi, G.; Koch, D. M.; Schmidt, G. A.; Unger, N.; Bauer, S. E. (2009). "Improved Attribution of Climate Forcing to Emissions". Science 326 (5953): 716–718. doi:10.1126/science.1174760. PMID 19900930. Bibcode: 2009Sci...326..716S. https://zenodo.org/record/1230902.
- ↑ Shindell, D. T.; Faluvegi, G.; Koch, D. M.; Schmidt, G. A.; Unger, N.; Bauer, S. E. (2009). "Improved Attribution of Climate Forcing to Emissions". Science 326 (5953): 716–728. doi:10.1126/science.1174760. PMID 19900930. Bibcode: 2009Sci...326..716S. https://zenodo.org/record/1230902.
- ↑ [1], https://clear.ucdavis.edu/explainers/biogenic-carbon-cycle-and-cattle.
- ↑ Food and Agriculture Organization of the United Nations (2013) "FAO Statistical Yearbook 2013 World Food and Agriculture". See data in Table 49 on p. 254.
- ↑ Inventory of U.S. Greenhouse Gas Emissions and Sinks: 1990–2014. 2016. https://www.epa.gov/ghgemissions/inventory-us-greenhouse-gas-emissions-and-sinks-1990-2014.
- ↑ Ripple, William J.; Pete Smith; Helmut Haberl; Stephen A. Montzka; Clive McAlpine & Douglas H. Boucher. 2014. "Ruminants, climate change and climate policy". Nature Climate Change. Volume 4 No. 1. pp. 2–5.
- ↑ Cicerone, R. J., and R. S. Oremland. 1988 "Biogeochemical Aspects of Atmospheric Methane"
- ↑ Yavitt, J. B. 1992. Methane, biogeochemical cycle. pp. 197–207 in Encyclopedia of Earth System Science, Vol. 3. Acad.Press, London.
- ↑ Bureau of Sport Fisheries and Wildlife (January 1965). "The American Buffalo". Conservation Note 12.
External links
| Wikisource has the text of the 1905 New International Encyclopedia article Ruminant. |
- Digestive Physiology of Herbivores – Colorado State University (Last updated on 13 July 2006)
- Britannica, The Editors of Encyclopaedia. "Ruminant". Encyclopædia Britannica, Invalid Date, https://www.britannica.com/animal/ruminant. Accessed 22 February 2021.
Wikidata ☰ Q192164 entry
 |



