Similarity measure
In statistics and related fields, a similarity measure or similarity function or similarity metric is a real-valued function that quantifies the similarity between two objects. Although no single definition of a similarity exists, usually such measures are in some sense the inverse of distance metrics: they take on large values for similar objects and either zero or a negative value for very dissimilar objects. Though, in more broad terms, a similarity function may also satisfy metric axioms.
Cosine similarity is a commonly used similarity measure for real-valued vectors, used in (among other fields) information retrieval to score the similarity of documents in the vector space model. In machine learning, common kernel functions such as the RBF kernel can be viewed as similarity functions.[1]
Use of different similarity measure formulas
Different types of similarity measures exist for various types of objects, depending on the objects being compared. For each type of object there are various similarity measurement formulas.[2]
Similarity between two data points
There are many various options available when it comes to finding similarity between two data points, some of which are a combination of other similarity methods. Some of the methods for similarity measures between two data points include Euclidean distance, Manhattan distance, Minkowski distance, and Chebyshev distance. The Euclidean distance formula is used to find the distance between two points on a plane, which is visualized in the image below. Manhattan distance is commonly used in GPS applications, as it can be used to find the shortest route between two addresses.[citation needed] When you generalize the Euclidean distance formula and Manhattan distance formula you are left with the Minkowski distance formulas, which can be used in a wide variety of applications.
- Euclidean distance
- Manhattan distance
- Minkowski distance
- Chebyshev distance
Similarity between strings
For comparing strings, there are various measures of string similarity that can be used. Some of these methods include edit distance, Levenshtein distance, Hamming distance, and Jaro distance. The best-fit formula is dependent on the requirements of the application. For example, edit distance is frequently used for natural language processing applications and features, such as spell-checking. Jaro distance is commonly used in record linkage to compare first and last names to other sources.
- Edit distance
- Levenshtein distance
- Lee distance
- Hamming distance
- Jaro distance
Similarity between two probability distributions
Typical measures of similarity for probability distributions are the Bhattacharyya distance and the Hellinger distance. Both provide a quantification of similarity for two probability distributions on the same domain, and they are mathematically closely linked. The Bhattacharyya distance does not fulfill the triangle inequality, meaning it does not form a metric. The Hellinger distance does form a metric on the space of probability distributions.
Similarity between two sets
The Jaccard index formula measures the similarity between two sets based on the number of items that are present in both sets relative to the total number of items. It is commonly used in recommendation systems and social media analysis . The Sørensen–Dice coefficient also compares the number of items in both sets to the total number of items present but the weight for the number of shared items is larger. The Sørensen–Dice coefficient is commonly used in biology applications, measuring the similarity between two sets of genes or species{{Citation needed|date=June 2024}
Similarity between two sequences
When comparing temporal sequences (time series), some similarity measures must additionally account for similarity of two sequences that are not fully aligned.
Use in clustering
Clustering or Cluster analysis is a data mining technique that is used to discover patterns in data by grouping similar objects together. It involves partitioning a set of data points into groups or clusters based on their similarities. One of the fundamental aspects of clustering is how to measure similarity between data points.
Similarity measures play a crucial role in many clustering techniques, as they are used to determine how closely related two data points are and whether they should be grouped together in the same cluster. A similarity measure can take many different forms depending on the type of data being clustered and the specific problem being solved.
One of the most commonly used similarity measures is the Euclidean distance, which is used in many clustering techniques including K-means clustering and Hierarchical clustering. The Euclidean distance is a measure of the straight-line distance between two points in a high-dimensional space. It is calculated as the square root of the sum of the squared differences between the corresponding coordinates of the two points. For example, if we have two data points and , the Euclidean distance between them is .
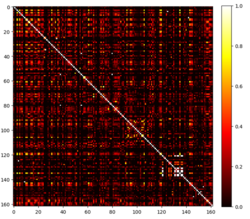
Another commonly used similarity measure is the Jaccard index or Jaccard similarity, which is used in clustering techniques that work with binary data such as presence/absence data [3] or Boolean data; The Jaccard similarity is particularly useful for clustering techniques that work with text data, where it can be used to identify clusters of similar documents based on their shared features or keywords.[4] It is calculated as the size of the intersection of two sets divided by the size of the union of the two sets: .
Similarities among 162 Relevant Nuclear Profile are tested using the Jaccard Similarity measure (see figure with heatmap). The Jaccard similarity of the nuclear profile ranges from 0 to 1, with 0 indicating no similarity between the two sets and 1 indicating perfect similarity with the aim of clustering the most similar nuclear profile.
Manhattan distance, also known as Taxicab geometry, is a commonly used similarity measure in clustering techniques that work with continuous data. It is a measure of the distance between two data points in a high-dimensional space, calculated as the sum of the absolute differences between the corresponding coordinates of the two points .
When dealing with mixed-type data, including nominal, ordinal, and numerical attributes per object, Gower's distance (or similarity) is a common choice as it can handle different types of variables implicitly. It first computes similarities between the pair of variables in each object, and then combines those similarities to a single weighted average per object-pair. As such, for two objects and having descriptors, the similarity is defined as: where the are non-negative weights and is the similarity between the two objects regarding their -th variable.
In spectral clustering, a similarity, or affinity, measure is used to transform data to overcome difficulties related to lack of convexity in the shape of the data distribution.[5] The measure gives rise to an -sized similarity matrix for a set of n points, where the entry in the matrix can be simply the (reciprocal of the) Euclidean distance between and , or it can be a more complex measure of distance such as the Gaussian .[5] Further modifying this result with network analysis techniques is also common.[6]
The choice of similarity measure depends on the type of data being clustered and the specific problem being solved. For example, working with continuous data such as gene expression data, the Euclidean distance or cosine similarity may be appropriate. If working with binary data such as the presence of a genomic loci in a nuclear profile, the Jaccard index may be more appropriate. Lastly, working with data that is arranged in a grid or lattice structure, such as image or signal processing data, the Manhattan distance is particularly useful for the clustering.
Use in recommender systems
Similarity measures are used to develop recommender systems. It observes a user's perception and liking of multiple items. On recommender systems, the method is using a distance calculation such as Euclidean Distance or Cosine Similarity to generate a similarity matrix with values representing the similarity of any pair of targets. Then, by analyzing and comparing the values in the matrix, it is possible to match two targets to a user's preference or link users based on their marks. In this system, it is relevant to observe the value itself and the absolute distance between two values.[7] Gathering this data can indicate a mark's likeliness to a user as well as how mutually closely two marks are either rejected or accepted. It is possible then to recommend to a user targets with high similarity to the user's likes.
Recommender systems are observed in multiple online entertainment platforms, in social media and streaming websites. The logic for the construction of this systems is based on similarity measures.[citation needed]
Use in sequence alignment
Similarity matrices are used in sequence alignment. Higher scores are given to more-similar characters, and lower or negative scores for dissimilar characters.
Nucleotide similarity matrices are used to align nucleic acid sequences. Because there are only four nucleotides commonly found in DNA (Adenine (A), Cytosine (C), Guanine (G) and Thymine (T)), nucleotide similarity matrices are much simpler than protein similarity matrices. For example, a simple matrix will assign identical bases a score of +1 and non-identical bases a score of −1. A more complicated matrix would give a higher score to transitions (changes from a pyrimidine such as C or T to another pyrimidine, or from a purine such as A or G to another purine) than to transversions (from a pyrimidine to a purine or vice versa). The match/mismatch ratio of the matrix sets the target evolutionary distance.[8][9] The +1/−3 DNA matrix used by BLASTN is best suited for finding matches between sequences that are 99% identical; a +1/−1 (or +4/−4) matrix is much more suited to sequences with about 70% similarity. Matrices for lower similarity sequences require longer sequence alignments.
Amino acid similarity matrices are more complicated, because there are 20 amino acids coded for by the genetic code, and so a larger number of possible substitutions. Therefore, the similarity matrix for amino acids contains 400 entries (although it is usually symmetric). The first approach scored all amino acid changes equally. A later refinement was to determine amino acid similarities based on how many base changes were required to change a codon to code for that amino acid. This model is better, but it doesn't take into account the selective pressure of amino acid changes. Better models took into account the chemical properties of amino acids.
One approach has been to empirically generate the similarity matrices. The Dayhoff method used phylogenetic trees and sequences taken from species on the tree. This approach has given rise to the PAM series of matrices. PAM matrices are labelled based on how many nucleotide changes have occurred, per 100 amino acids. While the PAM matrices benefit from having a well understood evolutionary model, they are most useful at short evolutionary distances (PAM10–PAM120). At long evolutionary distances, for example PAM250 or 20% identity, it has been shown that the BLOSUM matrices are much more effective.
The BLOSUM series were generated by comparing a number of divergent sequences. The BLOSUM series are labeled based on how much entropy remains unmutated between all sequences, so a lower BLOSUM number corresponds to a higher PAM number.
Use in computer vision
See also
- Affinity propagation – Algorithm in data mining
- Latent space – Embedding of data within a manifold based on a similarity function
- Similarity learning – Supervised learning of a similarity function
- Self-similarity matrix
- Semantic similarity – Natural language processing
- Similarity (network science)
- Statistical distance – Distance between two statistical objects
- String metric
- Similarity search – Searching for similar items in a data set
- Tf–idf
- Recurrence plot, a visualization tool of recurrences in dynamical (and other) systems
References
- ↑ Vert, Jean-Philippe; Tsuda, Koji; Schölkopf, Bernhard (2004). "A primer on kernel methods". Kernel Methods in Computational Biology. http://www.eecis.udel.edu/~lliao/cis841s06/primer_on_kernel_methods_vert.pdf.
- ↑ "Different Types of Similarity measurements". OpenGenus. https://iq.opengenus.org/similarity-measurements/.
- ↑ Chung, Neo Christopher; Miasojedow, BłaŻej; Startek, Michał; Gambin, Anna (2019). "Jaccard/Tanimoto similarity test and estimation methods for biological presence-absence data" (in en). BMC Bioinformatics 20 (S15): 644. doi:10.1186/s12859-019-3118-5. ISSN 1471-2105. PMID 31874610.
- ↑ International MultiConference of Engineers and Computer Scientists : IMECS 2013 : 13-15 March, 2013, the Royal Garden Hotel, Kowloon, Hong Kong. S. I. Ao, International Association of Engineers. Hong Kong: Newswood Ltd.. 2013. ISBN 978-988-19251-8-3. OCLC 842831996.
- ↑ 5.0 5.1 Ng, A.Y.; Jordan, M.I.; Weiss, Y. (2001), "On Spectral Clustering: Analysis and an Algorithm", Advances in Neural Information Processing Systems (MIT Press) 14: 849–856, https://www.researchgate.net/publication/320413363
- ↑ Li, Xin-Ye; Guo, Li-Jie (2012), "Constructing affinity matrix in spectral clustering based on neighbor propagation", Neurocomputing 97: 125–130, doi:10.1016/j.neucom.2012.06.023
- ↑ Bondarenko, Kirill (2019), Similarity metrics in recommender systems, https://bond-kirill-alexandrovich.medium.com/similarity-metrics-in-recommender-systems-aed9d3b2315f, retrieved 25 April 2023
- ↑ States, D; Gish, W; Altschul, S (1991). "Improved sensitivity of nucleic acid database searches using application-specific scoring matrices". Methods: A Companion to Methods in Enzymology 3 (1): 66. doi:10.1016/S1046-2023(05)80165-3.
- ↑ Sean R. Eddy (2004). "Where did the BLOSUM62 alignment score matrix come from?". Nature Biotechnology 22 (8): 1035–6. doi:10.1038/nbt0804-1035. PMID 15286655. http://informatics.umdnj.edu/bioinformatics/courses/5020/notes/BLOSUM62%20primer.pdf.
- ↑ Cite error: Invalid
<ref>tag; no text was provided for refs namedShapiro2001 - ↑ Eidenberger, Horst (2011). "Fundamental Media Understanding", atpress. ISBN 978-3-8423-7917-6.
- F. Gregory Ashby; Daniel M. Ennis (2007). "Similarity measures". Scholarpedia 2 (12): 4116. doi:10.4249/scholarpedia.4116. Bibcode: 2007SchpJ...2.4116A.
 |
