Biology:Allantoicase
| allantoicase | |||||||||
|---|---|---|---|---|---|---|---|---|---|
 Allantoicase homohexamer, Saccharomyces cerevisiae | |||||||||
| Identifiers | |||||||||
| EC number | 3.5.3.4 | ||||||||
| CAS number | 9025-21-2 | ||||||||
| Databases | |||||||||
| IntEnz | IntEnz view | ||||||||
| BRENDA | BRENDA entry | ||||||||
| ExPASy | NiceZyme view | ||||||||
| KEGG | KEGG entry | ||||||||
| MetaCyc | metabolic pathway | ||||||||
| PRIAM | profile | ||||||||
| PDB structures | RCSB PDB PDBe PDBsum | ||||||||
| Gene Ontology | AmiGO / QuickGO | ||||||||
| |||||||||
| Allantoicase | |||||||||
|---|---|---|---|---|---|---|---|---|---|
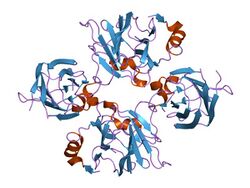 structure of allantoicase | |||||||||
| Identifiers | |||||||||
| Symbol | Allantoicase | ||||||||
| Pfam | PF03561 | ||||||||
| Pfam clan | CL0202 | ||||||||
| InterPro | IPR015908 | ||||||||
| SCOP2 | 1sg3 / SCOPe / SUPFAM | ||||||||
| |||||||||
Allantoicase is an enzyme (EC 3.5.3.4) that in humans is encoded by the ALLC gene. Allantoicase catalyzes the chemical reaction
- allantoate + H2O [math]\displaystyle{ \rightleftharpoons }[/math] (S)-ureidoglycolate + urea
Thus, the two substrates of this enzyme are allantoate and H2O, whereas its two products are (S)-ureidoglycolate and urea.
This enzyme belongs to the family of hydrolases, those acting on carbon-nitrogen bonds other than peptide bonds, specifically in linear amidines. The systematic name of this enzyme class is allantoate amidinohydrolase. This enzyme participates in purine metabolism by facilitating the utilization of purines as secondary nitrogen sources under nitrogen-limiting conditions. While purine degradation converges to uric acid in all vertebrates, its further degradation varies from species to species. Uric acid is excreted by birds, reptiles, and some mammals that do not have a functional uricase gene, whereas other mammals produce allantoin. Amphibians and microorganisms produce ammonia and carbon dioxide using the uricolytic pathway. Allantoicase performs the second step in this pathway catalyzing the conversion of allantoate into ureidoglycolate and urea.
Structural studies
As of late 2007, two structures have been solved for this class of enzymes, with PDB accession codes 1O59 and 1SG3.
The structure of allantoicase is best described as being composed of two repeats (the allantoicase repeats: AR1 and AR2), which are connected by a flexible linker. The crystal structure, resolved at 2.4A resolution, reveals that AR1 has a very similar fold to AR2, both repeats being jelly-roll motifs, composed of four-stranded and five-stranded antiparallel beta-sheets.[1] Each jelly-roll motif has two conserved surface patches that probably constitute the active site.[2]
References
- ↑ "Crystal structure of an allantoicase (YIR029W) from Saccharomyces cerevisiae at 2.4 A resolution". Proteins 56 (3): 619–24. August 2004. doi:10.1002/prot.20164. PMID 15229895.
- ↑ "Crystal structure of yeast allantoicase reveals a repeated jelly roll motif". The Journal of Biological Chemistry 279 (22): 23447–52. May 2004. doi:10.1074/jbc.M401336200. PMID 15020593.
Further reading
- "Microdosage photometrique de l'allantoine en solutions pures et dans l'urine". Enzymologia 9: 5–9. 1940.
- "Allantoicase and ureidoglycolase in Pseudomonas and Penicillium species". Biochimica et Biophysica Acta (BBA) - Enzymology and Biological Oxidation 118 (2): 387–95. May 1966. doi:10.1016/S0926-6593(66)80047-4. PMID 4960174.
- "Impact of hypothermia on the immunologic response after trauma and elective surgery". Surgical Technology International 14: 41–50. 2005. PMID 16525953.
- "Hydrolysis, racemization and absolute configuration of ureidoglycolate, a substrate of allantoicase". Biochimica et Biophysica Acta (BBA) - Enzymology 198 (3): 569–82. March 1970. doi:10.1016/0005-2744(70)90134-8. PMID 4314237.
 |

