Biology:Nitrilase
| nitrilase 1 | |
|---|---|
 | |
| Identifiers | |
| Symbol | NIT1 |
| NCBI gene | 4817 |
| HGNC | 7828 |
| OMIM | 604618 |
| PDB | 3IVZ |
| RefSeq | NM_005600 |
| UniProt | Q86X76 |
| Other data | |
| Locus | Chr. 1 pter-qter |
Nitrilase enzymes (nitrile aminohydrolase; EC 3.5.5.1) catalyse the hydrolysis of nitriles to carboxylic acids and ammonia, without the formation of "free" amide intermediates.[1] Nitrilases are involved in natural product biosynthesis and post translational modifications in plants, animals, fungi and certain prokaryotes. Nitrilases can also be used as catalysts in preparative organic chemistry. Among others, nitrilases have been used for the resolution of racemic mixtures. Nitrilase should not be confused with nitrile hydratase (nitrile hydro-lyase; EC 4.2.1.84) which hydrolyses nitriles to amides. Nitrile hydratases are almost invariably co-expressed with an amidase, which converts the amide to the carboxylic acid. Consequently, it can sometimes be difficult to distinguish nitrilase activity from nitrile hydratase plus amidase activity.
Mechanism
Nitrilase was first discovered in the early 1960s for its ability to catalyze the hydration of a nitrile to a carboxylic acid.[2] Although it was known at the time that nitrilase could operate with wide substrate specificity in producing the corresponding acid, later studies reported the first NHase (nitrile hydratase) activity exhibited by nitrilase.[3][4] That is, amide compounds could also be formed via nitrile hydrolysis. Further research has revealed several conditions that promote amide formation, which are outlined below.[4]
- Early release of the enzyme-bound substrate after the first water hydrolysis followed by delayed addition of the second water
- Low temperature and increased pH conditions. For bioconversions by nitrilase for most bacteria and fungi, the optimal pH range is between 7.0-8.0 and the optimal temperature range is between 30 and 50 °C.
- Electron withdrawing groups at the ⍺-position

Below is a list of steps involved in transforming a generic nitrile compound with nitrilase:[4]
- The electrophilic carbon of the nitrile is subject to nucleophilic attack by one of the two SH groups on nitrilase.
- The thioimidate formed is subsequently hydrolyzed to the acylenzyme and ammonia is created as a byproduct.
- The acylenzyme can undergo one of two pathways depending on the conditions highlighted above:
- Further hydrolyzation of the acylenzyme with water produces the carboxylic acid and the regenerated enzyme.
- The acylenzyme is hydrolyzed by ammonia, displacing the enzyme and forming the amide product.
Structure
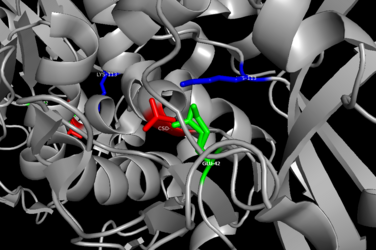
Most nitrilases are made up of a single polypeptide ranging from 32 to 45 kDa,[7] and its structure is an ⍺-β-β-⍺ fold.[4] The favored form of the enzyme is a large filament consisting of 6-26 subunits.[7] Nitrilase exploits the Lys-Cys-Glu catalytic triad which is essential for its active site function and enhancing its performance.[4][7]
The structure of a thermoactive nitrilase from P. abyssi consists of a 2-fold symmetric dimer in which each subunit contains 262 residues.[8][9] Similar to other nitrilases in the nitrilase family, each subunit has an ⍺-β-β-⍺ sandwich fold; when the two subunits come together and interact, the protein forms a ‘super-sandwich’ (⍺-β-β-⍺-⍺-β-β-⍺) structure.[6] In order to dimerize, the C-terminals of each subunit extend out from the core and interact with each other, and this is largely made possible by the salt bridges formed between arginine and glutamate residues.[6]
Although the exact binding mechanism to the nitrile substrate still remains unknown, by drawing comparisons between the sequence and structure with other nitrilases, the catalytic triad was determined to consist of Glu 42, Lys 113, and Cys 146.[6][4][7] With the aid of protein modeling programs, Glu 42 was observed to be the catalytic base in activating the nucleophile (Cys 146) based on the relatively short distance between the O in Glu and S in Cys. Likewise, Lys 113 was inferred to be the catalytic acid responsible for proton transfer to the substrate.[8][10]
Biological Function
Nitrilases have critical roles in plant-microbe interactions for defense, detoxification, nitrogen utilization, and plant hormone synthesis.[11] In plants, there are two distinguishable groups in regard to substrate specificity: those with high hydrolytic activity towards arylacetonitriles and those with high activity towards β-cyano-L-alanine. NIT1, 2, and 3 of the A. thaliana species are examples of the first group of plant nitrilases (arylacetonitrilases) which hydrolyze the nitriles produced during the synthesis or degradation of cyanogenic glycosides and glucosinolates. The arylcetonitrile substrates for these particular enzymes consist of phenylpropionitrile and other products that result from glucosinolate metabolism.[11][12] NIT4 however, belongs to the second group of plant nitrilases and is critical for cyanide detoxification in plants.[3][11][13]
Moreover, microbes could also potentially utilize nitrilase for detoxifying and assimilating nitriles and cyanide that exist in the plant environment.[11] An example of this is the β-cyano-L-alanine nitrilase by the plant bacterium P. fluorescens SBW25.[14] Although it is unknown whether this plant bacterium encounters toxic levels of β-cyano-ʟ-alanine in natural settings, nitrilase activity has been observed in cyanogenic plants; thus, it seems that the nitrilase serves as a predominant mechanism for detoxifying cyanide instead of β-cyano-ʟ-alanine.[11][14] Other bacterial applications of nitrilases produced by plant-associated microorganisms include the degradation of plant nitriles for a carbon and nitrogen source. P. fluorescens EBC191 hydrolyzes many arylacetonitriles, namely mandelonitrile, which serves as a defense against herbivores.[11][15][16]
Further reading
- "Enzyme stabilizer DTT catalyzes nitrilase analogue hydrolysis of nitriles". Chemical Communications (12): 1298–300. March 2006. doi:10.1039/B516937B. PMID 16538253.
References
- ↑ 1.0 1.1 "The nitrilase superfamily: classification, structure and function". Genome Biology 2 (1): REVIEWS0001. 2001. doi:10.1186/gb-2001-2-1-reviews0001. PMID 11380987.
- ↑ "Nitrilase. I. Occurrence, Preparation, and General Properties of the Enzyme". Archives of Biochemistry and Biophysics 105: 133–41. April 1964. doi:10.1016/0003-9861(64)90244-9. PMID 14165487.
- ↑ 3.0 3.1 "The Arabidopsis thaliana isogene NIT4 and its orthologs in tobacco encode beta-cyano-L-alanine hydratase/nitrilase". The Journal of Biological Chemistry 276 (4): 2616–21. January 2001. doi:10.1074/jbc.M007890200. PMID 11060302.
- ↑ 4.0 4.1 4.2 4.3 4.4 4.5 "Nitrilases in nitrile biocatalysis: recent progress and forthcoming research". Microbial Cell Factories 11: 142. October 2012. doi:10.1186/1475-2859-11-142. PMID 23106943.
- ↑ "Cloning of a nitrilase gene from the cyanobacterium Synechocystis sp. strain PCC6803 and heterologous expression and characterization of the encoded protein". Applied and Environmental Microbiology 69 (8): 4359–66. 2003. doi:10.1128/AEM.69.8.4359-4366.2003. PMID 12902216.
- ↑ 6.0 6.1 6.2 6.3 "Crystallographic analysis of a thermoactive nitrilase". Journal of Structural Biology 173 (2): 294–302. February 2011. doi:10.1016/j.jsb.2010.11.017. PMID 21095228.
- ↑ 7.0 7.1 7.2 7.3 "The nitrilase family of CN hydrolysing enzymes - a comparative study". Journal of Applied Microbiology 95 (6): 1161–74. 2003. doi:10.1046/j.1365-2672.2003.02123.x. PMID 14632988.
- ↑ 8.0 8.1 "Crystal structure of N-carbamyl-D-amino acid amidohydrolase with a novel catalytic framework common to amidohydrolases". Structure 8 (7): 729–37. July 2000. doi:10.1016/s0969-2126(00)00160-x. PMID 10903946.
- ↑ "Crystal structure of the worm NitFhit Rosetta Stone protein reveals a Nit tetramer binding two Fhit dimers". Current Biology 10 (15): 907–17. 2017-07-27. doi:10.1016/s0960-9822(00)00621-7. PMID 10959838.
- ↑ "Detecting and overcoming crystal twinning". Macromolecular Crystallography Part A. Methods in Enzymology. 276. 1997-01-01. pp. 344–58. doi:10.1016/s0076-6879(97)76068-3. ISBN 9780121821777.
- ↑ 11.0 11.1 11.2 11.3 11.4 11.5 "Nitrilase enzymes and their role in plant-microbe interactions". Microbial Biotechnology 2 (4): 441–51. July 2009. doi:10.1111/j.1751-7915.2009.00111.x. PMID 21255276.
- ↑ "Enzymatic characterization of the recombinant Arabidopsis thaliana nitrilase subfamily encoded by the NIT2/NIT1/NIT3-gene cluster". Planta 212 (4): 508–16. March 2001. doi:10.1007/s004250000420. PMID 11525507.
- ↑ "Primary or secondary? Versatile nitrilases in plant metabolism". Phytochemistry 69 (15): 2655–67. November 2008. doi:10.1016/j.phytochem.2008.08.020. PMID 18842274.
- ↑ 14.0 14.1 "A conserved mechanism for nitrile metabolism in bacteria and plants". The Plant Journal 57 (2): 243–53. January 2009. doi:10.1111/j.1365-313X.2008.03682.x. PMID 18786181.
- ↑ "Nitrilase from Pseudomonas fluorescens EBC191: cloning and heterologous expression of the gene and biochemical characterization of the recombinant enzyme". Microbiology 151 (Pt 11): 3639–48. November 2005. doi:10.1099/mic.0.28246-0. PMID 16272385.
- ↑ "Natural nitriles and their metabolism". World Journal of Microbiology & Biotechnology 6 (2): 83–108. June 1990. doi:10.1007/BF01200927. PMID 24429979.
External links
- nitrilase at the US National Library of Medicine Medical Subject Headings (MeSH)
 |
