Biology:Peptide deformylase
| Peptide deformylase | |||||||||
|---|---|---|---|---|---|---|---|---|---|
 Escherichia coli peptide deformylase structure | |||||||||
| Identifiers | |||||||||
| EC number | 3.5.1.88 | ||||||||
| Databases | |||||||||
| IntEnz | IntEnz view | ||||||||
| BRENDA | BRENDA entry | ||||||||
| ExPASy | NiceZyme view | ||||||||
| KEGG | KEGG entry | ||||||||
| MetaCyc | metabolic pathway | ||||||||
| PRIAM | profile | ||||||||
| PDB structures | RCSB PDB PDBe PDBsum | ||||||||
| Gene Ontology | AmiGO / QuickGO | ||||||||
| |||||||||
In enzymology, a peptide deformylase (EC 3.5.1.88) is an enzyme that removes the formyl group from the N terminus of nascent polypeptide chains in eubacteria, mitochondria and chloroplasts.[1]
Peptide deformylases are metaloenzymes monomers and bind a metal cofactor, typically Fe(II) or Zn, in an active site. Cofactor identity impacts catalytic efficiency.[2]
There are two types of peptide deformylases, types I and II, which differ in structure mainly in the outer surface of the protein.
Human gene PDF (gene) posesses this activity.
Function
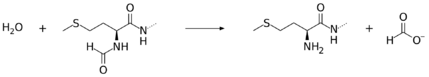
Peptide deformylase removes the formyl group from the N terminus of nascent polypeptides as they are synthesized by the ribosome. The function of peptide deformylase can be described by the following equation, where formyl-L-methionyl peptide and water react under the formation of formate and methionyl peptide:
- H2O + formyl-L-methionyl peptide [math]\displaystyle{ \rightleftharpoons }[/math] methionyl peptide + formate
This reaction takes place on the surface of the ribosome, where the C-terminal alpha-helix of the peptide deformylase interacts with a grove between ribosomal proteins uL22 and bL32, and rRNA.[3]
For its function this enzyme belongs to the family of hydrolases, those acting on carbon-nitrogen bonds other than peptide bonds, specifically in linear amides. The systematic name of this enzyme class is formyl-L-methionyl peptide amidohydrolase.
Structural studies
As of late 2007, 34 structures have been solved for this class of enzymes, with PDB accession codes 1IX1, 1LM4, 1LM6, 1LME, 1LQW, 1LQY, 1LRU, 1LRY, 1N5N, 1Q1Y, 1S17, 1SV2, 1SZZ, 1V3Y, 1VEV, 1VEY, 1VEZ, 1WS0, 1WS1, 1XEM, 1XEN, 1XEO, 1Y6H, 1ZXZ, 1ZY0, 1ZY1, 2AI7, 2AI8, 2AI9, 2AIA, 2AIE, 2EW5, 2EW6, and 2EW7.
See also
References
- ↑ "Structure and activity of human mitochondrial peptide deformylase, a novel cancer target". Journal of Molecular Biology 387 (5): 1211–1228. April 2009. doi:10.1016/j.jmb.2009.02.032. PMID 19236878.
- ↑ "Structure of peptide deformylase and identification of the substrate binding site". The Journal of Biological Chemistry 273 (19): 11413–11416. May 1998. doi:10.1074/jbc.273.19.11413. PMID 9565550.
- ↑ "Binding of the peptide deformylase on the ribosome surface modulates the exit tunnel interior". Biophysical Journal 121 (23): 4443–4451. December 2022. doi:10.1016/j.bpj.2022.11.004. PMID 36335428. Bibcode: 2022BpJ...121.4443M.
Further reading
- "On the release of the formyl group from nascent protein". Journal of Molecular Biology 33 (3): 571–589. May 1968. doi:10.1016/0022-2836(68)90307-0. PMID 4973445.
- "Genetic characterization of polypeptide deformylase, a distinctive enzyme of eubacterial translation". The EMBO Journal 13 (4): 914–923. February 1994. doi:10.1002/j.1460-2075.1994.tb06335.x. PMID 8112305.
- "Crystal structure of the Escherichia coli peptide deformylase". Biochemistry 36 (45): 13904–13909. November 1997. doi:10.1021/bi9711543. PMID 9374869.
- "Structure of peptide deformylase and identification of the substrate binding site". The Journal of Biological Chemistry 273 (19): 11413–11416. May 1998. doi:10.1074/jbc.273.19.11413. PMID 9565550.
- "Iron center, substrate recognition and mechanism of peptide deformylase". Nature Structural Biology 5 (12): 1053–1058. December 1998. doi:10.1038/4162. PMID 9846875.
- "Peptide deformylase: a new type of mononuclear iron protein". J. Am. Chem. Soc. 119 (50): 12418–12419. 1997. doi:10.1021/ja9734096.
- "Isolation and crystallization of functionally competent Escherichia coli peptide deformylase forms containing either iron or nickel in the active site". Biochemical and Biophysical Research Communications 246 (2): 342–346. May 1998. doi:10.1006/bbrc.1998.8616. PMID 9610360.
- "Characterization of cobalt(II)-substituted peptide deformylase: function of the metal ion and the catalytic residue Glu-133". Biochemistry 39 (4): 779–790. February 2000. doi:10.1021/bi9919899. PMID 10651644.
- "Determination of substrate specificity for peptide deformylase through the screening of a combinatorial peptide library". Biochemistry 38 (2): 643–650. January 1999. doi:10.1021/bi9820412. PMID 9888804.
- "Substrate recognition and selectivity of peptide deformylase. Similarities and differences with metzincins and thermolysin". Journal of Molecular Biology 289 (5): 1445–1457. June 1999. doi:10.1006/jmbi.1999.2832. PMID 10373378.
- "Peptide deformylase as a target for new generation, broad spectrum antimicrobial agents". Molecular Microbiology 36 (6): 1197–1205. June 2000. doi:10.1046/j.1365-2958.2000.01908.x. PMID 10931273.
- "Peptide deformylase: a target for novel antibiotics?". Expert Opinion on Therapeutic Targets 5 (1): 23–40. February 2001. doi:10.1517/14728222.5.1.23. PMID 15992166.
 |