Biology:Symbiogenesis
Symbiogenesis (endosymbiotic theory, or serial endosymbiotic theory[2]) is the leading evolutionary theory of the origin of eukaryotic cells from prokaryotic organisms.[3] The theory holds that mitochondria, plastids such as chloroplasts, and possibly other organelles of eukaryotic cells are descended from formerly free-living prokaryotes (more closely related to the Bacteria than to the Archaea) taken one inside the other in endosymbiosis. Mitochondria appear to be phylogenetically related to Rickettsiales bacteria, while chloroplasts are thought to be related to cyanobacteria.
The idea that chloroplasts were originally independent organisms that merged into a symbiotic relationship with other one-celled organisms dates back to the 19th century, when it was espoused by researchers such as Andreas Schimper. The endosymbiotic theory was articulated in 1905 and 1910 by the Russian botanist Konstantin Mereschkowski, and advanced and substantiated with microbiological evidence by Lynn Margulis in 1967.
Among the many lines of evidence supporting symbiogenesis are that new mitochondria and plastids are formed only by splitting in two, and that cells cannot create new ones otherwise; that the transport proteins called porins are found in the outer membranes of mitochondria, chloroplasts, and bacterial cell membranes; that cardiolipin is found only in the inner mitochondrial membrane and bacterial cell membranes; and that some mitochondria and plastids contain single circular DNA molecules similar to the circular chromosomes of bacteria.
History
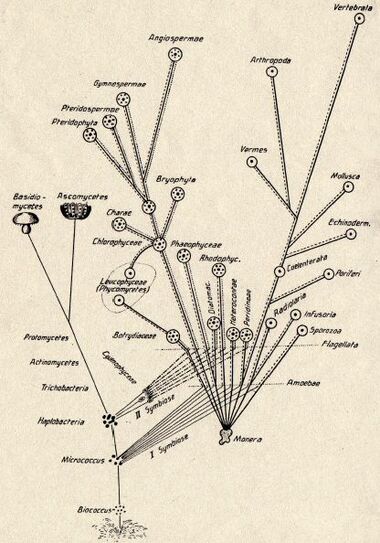
The Russian botanist Konstantin Mereschkowski first outlined the theory of symbiogenesis (from Greek: σύν syn "together", βίος bios "life", and γένεσις genesis "origin, birth") in his 1905 work, The nature and origins of chromatophores in the plant kingdom, and then elaborated it in his 1910 The Theory of Two Plasms as the Basis of Symbiogenesis, a New Study of the Origins of Organisms.[5][6][7] Mereschkowski proposed that complex life-forms had originated by two episodes of symbiogenesis, the incorporation of symbiotic bacteria to form successively nuclei and chloroplasts.[4] Mereschkowski knew of the work of botanist Andreas Schimper. In 1883, Schimper had observed that the division of chloroplasts in green plants closely resembled that of free-living cyanobacteria. Schimper had tentatively proposed (in a footnote) that green plants had arisen from a symbiotic union of two organisms.[8] In 1918 the French scientist Paul Jules Portier published Les Symbiotes, in which he claimed that the mitochondria originated from a symbiosis process.[9][10] Ivan Wallin advocated the idea of an endosymbiotic origin of mitochondria in the 1920s.[11][12] The Russian botanist Boris Kozo-Polyansky became the first to explain the theory in terms of Darwinian evolution.[13] In his 1924 book A New Principle of Biology. Essay on the Theory of Symbiogenesis,[14] he wrote, "The theory of symbiogenesis is a theory of selection relying on the phenomenon of symbiosis."[15]
These theories did not gain traction until more detailed electron-microscopic comparisons between cyanobacteria and chloroplasts were made, such as by Hans Ris in 1961 and 1962.[16][17] These, combined with the discovery that plastids and mitochondria contain their own DNA,[18] led to a resurrection of the idea of symbiogenesis in the 1960s. Lynn Margulis advanced and substantiated the theory with microbiological evidence in a 1967 paper, On the origin of mitosing cells.[19] In her 1981 work Symbiosis in Cell Evolution she argued that eukaryotic cells originated as communities of interacting entities, including endosymbiotic spirochaetes that developed into eukaryotic flagella and cilia. This last idea has not received much acceptance, because flagella lack DNA and do not show ultrastructural similarities to bacteria or to archaea (see also: Evolution of flagella and Prokaryotic cytoskeleton). According to Margulis and Dorion Sagan,[20] "Life did not take over the globe by combat, but by networking" (i.e., by cooperation). Christian de Duve proposed that the peroxisomes may have been the first endosymbionts, allowing cells to withstand growing amounts of free molecular oxygen in the Earth's atmosphere. However, it now appears that peroxisomes may be formed de novo, contradicting the idea that they have a symbiotic origin.[21] The fundamental theory of symbiogenesis as the origin of mitochondria and chloroplasts is now widely accepted.[3]
From endosymbionts to organelles
Biologists usually distinguish organelles from endosymbionts – whole organisms living inside other organisms – by their reduced genome sizes.[23] As an endosymbiont evolves into an organelle, most of its genes are transferred to the host cell genome.[24] The host cell and organelle therefore need to develop a transport mechanism that enables the return of the protein products needed by the organelle but now manufactured by the cell.[25]
Free-living ancestors
Alphaproteobacteria were formerly thought to be the free-living organisms most closely related to mitochondria.[25] Later research indicates that mitochondria are most closely related to Pelagibacterales bacteria, in particular, those in the SAR11 clade.[26][27]
Nitrogen-fixing filamentous cyanobacteria are the free-living organisms most closely related to plastids.[25][28][29]
Both cyanobacteria and alphaproteobacteria maintain a large (>6 Mb) genome encoding thousands of proteins.[25] Plastids and mitochondria exhibit a dramatic reduction in genome size when compared with their bacterial relatives.[25] Chloroplast genomes in photosynthetic organisms are normally 120–200 kb[30] encoding 20–200 proteins[25] and mitochondrial genomes in humans are approximately 16 kb and encode 37 genes, 13 of which are proteins.[31] Using the example of the freshwater amoeboid, however, Paulinella chromatophora, which contains chromatophores found to be evolved from cyanobacteria, Keeling and Archibald argue that this is not the only possible criterion; another is that the host cell has assumed control of the regulation of the former endosymbiont's division, thereby synchronizing it with the cell's own division.[23] Nowack and her colleagues gene sequenced the chromatophore (1.02 Mb) and found that only 867 proteins were encoded by these photosynthetic cells. Comparisons with their closest free living cyanobacteria of the genus Synechococcus (having a genome size 3 Mb, with 3300 genes) revealed that chromatophores had undergone a drastic genome shrinkage. Chromatophores contained genes that were accountable for photosynthesis but were deficient in genes that could carry out other biosynthetic functions; this observation suggests that these endosymbiotic cells are highly dependent on their hosts for their survival and growth mechanisms. Thus, these chromatophores were found to be non-functional for organelle-specific purposes when compared with mitochondria and plastids. This distinction could have promoted the early evolution of photosynthetic organelles.[32]
The loss of genetic autonomy, that is, the loss of many genes from endosymbionts, occurred very early in evolutionary time.[33] Taking into account the entire original endosymbiont genome, there are three main possible fates for genes over evolutionary time. The first is the loss of functionally redundant genes,[33] in which genes that are already represented in the nucleus are eventually lost. The second is the transfer of genes to the nucleus, while the third is that genes remain in the organelle that was once an organism.[25][33][34][35][36] The loss of autonomy and integration of the endosymbiont with its host can be primarily attributed to nuclear gene transfer.[36] As organelle genomes have been greatly reduced over evolutionary time, nuclear genes have expanded and become more complex.[25] As a result, many plastid and mitochondrial processes are driven by nuclear encoded gene products.[25] In addition, many nuclear genes originating from endosymbionts have acquired novel functions unrelated to their organelles.[25][36]
Gene transfer mechanisms
The mechanisms of gene transfer are not fully known; however, multiple hypotheses exist to explain this phenomenon. The possible mechanisms include the Complementary DNA (cDNA) hypothesis and the bulk flow hypothesis.[25][34]
The cDNA hypothesis involves the use of messenger RNA (mRNAs) to transport genes from organelles to the nucleus where they are converted to cDNA and incorporated into the genome.[25][34] The cDNA hypothesis is based on studies of the genomes of flowering plants. Protein coding RNAs in mitochondria are spliced and edited using organelle-specific splice and editing sites. Nuclear copies of some mitochondrial genes, however, do not contain organelle-specific splice sites, suggesting a processed mRNA intermediate. The cDNA hypothesis has since been revised as edited mitochondrial cDNAs are unlikely to recombine with the nuclear genome and are more likely to recombine with their native mitochondrial genome. If the edited mitochondrial sequence recombines with the mitochondrial genome, mitochondrial splice sites would no longer exist in the mitochondrial genome. Any subsequent nuclear gene transfer would therefore also lack mitochondrial splice sites.[25]
The bulk flow hypothesis is the alternative to the cDNA hypothesis, stating that escaped DNA, rather than mRNA, is the mechanism of gene transfer.[25][34] According to this hypothesis, disturbances to organelles, including autophagy (normal cell destruction), gametogenesis (the formation of gametes), and cell stress release DNA which is imported into the nucleus and incorporated into the nuclear DNA using non-homologous end joining (repair of double stranded breaks).[34] For example, in the initial stages of endosymbiosis, due to a lack of major gene transfer, the host cell had little to no control over the endosymbiont. The endosymbiont underwent cell division independently of the host cell, resulting in many "copies" of the endosymbiont within the host cell. Some of the endosymbionts lysed (burst), and high levels of DNA were incorporated into the nucleus. A similar mechanism is thought to occur in tobacco plants, which show a high rate of gene transfer and whose cells contain multiple chloroplasts.[33] In addition, the bulk flow hypothesis is also supported by the presence of non-random clusters of organelle genes, suggesting the simultaneous movement of multiple genes.[34]
Ford Doolittle proposed that (whatever the mechanism) gene transfer behaves like a ratchet, resulting in unidirectional transfer of genes from the organelle to the nuclear genome.[37] When genetic material from an organelle is incorporated into the nuclear genome, either the organelle or nuclear copy of the gene may be lost from the population. If the organelle copy is lost and this is fixed, or lost through genetic drift, a gene is successfully transferred to the nucleus. If the nuclear copy is lost, horizontal gene transfer can occur again, and the cell can ‘try again’ to have successful transfer of genes to the nucleus.[37] In this ratchet-like way, genes from an organelle would be expected to accumulate in the nuclear genome over evolutionary time.[37]
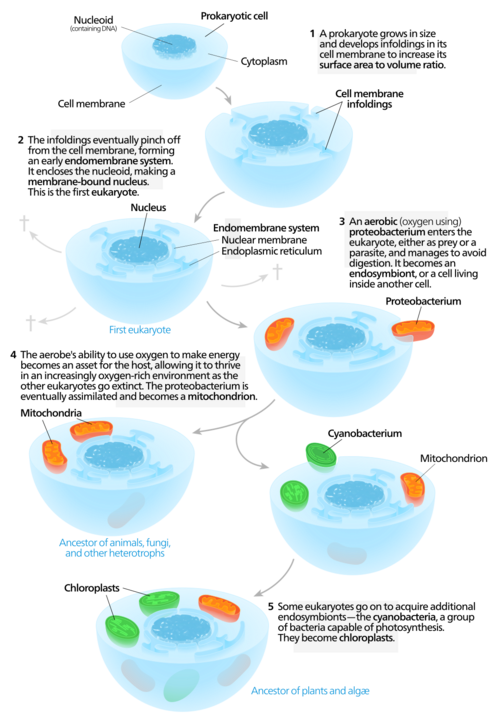
Endosymbiosis of protomitochondria
Endosymbiotic theory for the origin of mitochondria suggests that the proto-eukaryote engulfed a protomitochondrion, and this endosymbiont became an organelle, a major step in eukaryogenesis, the creation of the eukaryotes.[38]
Mitochondria

Mitochondria are organelles that synthesize the energy-carrying molecule ATP for the cell by metabolizing carbon-based macromolecules.[39] The presence of DNA in mitochondria and proteins, derived from mtDNA, suggest that this organelle may have been a prokaryote prior to its integration into the proto-eukaryote.[40] Mitochondria are regarded as organelles rather than endosymbionts because mitochondria and the host cells share some parts of their genome, undergo division simultaneously, and provide each other with means to produce energy.[40] The endomembrane system and nuclear membrane were hypothesized to have derived from the protomitochondria.[41][42][43]
Nuclear membrane
The presence of a nucleus is one major difference between eukaryotes and prokaryotes.[44] Some conserved nuclear proteins between eukaryotes and prokaryotes suggest that these two types had a common ancestor.[45] Another theory behind nucleation is that early nuclear membrane proteins caused the cell membrane to fold and form a sphere with pores like the nuclear envelope.[46] As a way of forming a nuclear membrane, endosymbiosis could be expected to use less energy than if the cell was to develop a metabolic process to fold the cell membrane for the purpose.[42] Digesting engulfed cells without energy-producing mitochondria would have been challenging for the host cell.[41] On this view, membrane-bound bubbles or vesicles leaving the protomitochondria may have formed the nuclear envelope.[41]
The process of symbiogenesis by which the early eukaryotic cell integrated the proto-mitochondrion likely included protection of the archaeal host genome from the release of reactive oxygen species. These would have been formed during oxidative phosphorylation and ATP production by the proto-mitochondrion. The nuclear membrane may have evolved as an adaptive innovation for protecting against nuclear genome DNA damage caused by reactive oxygen species.[47] Substantial transfer of genes from the ancestral proto-mitochondrial genome to the nuclear genome likely occurred during early eukaryotic evolution.[48] The greater protection of the nuclear genome against reactive oxygen species afforded by the nuclear membrane may explain the adaptive benefit of this gene transfer.
Endomembrane system
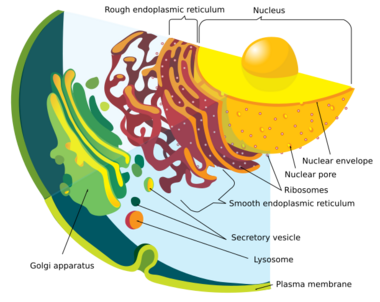
Modern eukaryotic cells use the endomembrane system to transport products and wastes in, within, and out of cells. The membrane of nuclear envelope and endomembrane vesicles are composed of similar membrane proteins.[49] These vesicles also share similar membrane proteins with the organelle they originated from or are traveling towards.[50] This suggests that what formed the nuclear membrane also formed the endomembrane system. Prokaryotes do not have a complex internal membrane network like eukaryotes, but they could produce extracellular vesicles from their outer membrane.[41] After the early prokaryote was consumed by a proto-eukaryote, the prokaryote would have continued to produce vesicles that accumulated within the cell.[41] Interaction of internal components of vesicles may have led to the endoplasmic reticulum and the Golgi apparatus, both being parts of the endomembrane system.[41]
Cytoplasm
The syntrophy hypothesis, proposed by López-García and Moreira around the year 2000, suggested that eukaryotes arose by combining the metabolic capabilities of an archaean, a fermenting deltaproteobacterium, and a methanotrophic alphaproteobacterium which became the mitochondrion. In 2020, the same team updated their syntrophy proposal to cover an Asgard archaean that produced hydrogen with deltaproteobacterium that oxidised sulphur. A third organism, an alphaproteobacterium able to respire both aerobically and anaerobically, and to oxidise sulphur, developed into the mitochondrion; it may possibly also have been able to photosynthesise.[51]
Organellar genomes
Plastomes and mitogenomes

Some endosymbiont genes remain in the organelles. Plastids and mitochondria retain genes encoding rRNAs, tRNAs, proteins involved in redox reactions, and proteins required for transcription, translation, and replication. There are many hypotheses to explain why organelles retain a small portion of their genome; however no one hypothesis will apply to all organisms, and the topic is still quite controversial. The hydrophobicity hypothesis states that highly hydrophobic (water hating) proteins (such as the membrane bound proteins involved in redox reactions) are not easily transported through the cytosol and therefore these proteins must be encoded in their respective organelles. The code disparity hypothesis states that the limit on transfer is due to differing genetic codes and RNA editing between the organelle and the nucleus. The redox control hypothesis states that genes encoding redox reaction proteins are retained in order to effectively couple the need for repair and the synthesis of these proteins. For example, if one of the photosystems is lost from the plastid, the intermediate electron carriers may lose or gain too many electrons, signalling the need for repair of a photosystem. The time delay involved in signalling the nucleus and transporting a cytosolic protein to the organelle results in the production of damaging reactive oxygen species. The final hypothesis states that the assembly of membrane proteins, particularly those involved in redox reactions, requires coordinated synthesis and assembly of subunits; however, translation and protein transport coordination is more difficult to control in the cytoplasm.[25][30][33][52]
Non-photosynthetic plastid genomes
The majority of the genes in the mitochondria and plastids are related to the expression (transcription, translation and replication) of genes encoding proteins involved in either photosynthesis (in plastids) or cellular respiration (in mitochondria). One might predict that the loss of photosynthesis or cellular respiration would allow for the complete loss of the plastid genome or the mitochondrial genome respectively.[25][30][33] While there are numerous examples of mitochondrial descendants (mitosomes and hydrogenosomes) that have lost their entire organellar genome,[50] non-photosynthetic plastids tend to retain a small genome. There are two main hypotheses to explain this occurrence:[33][53]
The essential tRNA hypothesis notes that there have been no documented functional plastid-to-nucleus gene transfers of genes encoding RNA products (tRNAs and rRNAs). As a result, plastids must make their own functional RNAs or import nuclear counterparts. The genes encoding tRNA-Glu and tRNA-fmet, however, appear to be indispensable. The plastid is responsible for haem biosynthesis, which requires plastid encoded tRNA-Glu (from the gene trnE) as a precursor molecule. Like other genes encoding RNAs, trnE cannot be transferred to the nucleus. In addition, it is unlikely trnE could be replaced by a cytosolic tRNA-Glu as trnE is highly conserved; single base changes in trnE have resulted in the loss of haem synthesis. The gene for tRNA-formylmethionine (tRNA-fmet) is also encoded in the plastid genome and is required for translation initiation in both plastids and mitochondria. A plastid is required to continue expressing the gene for tRNA-fmet so long as the mitochondrion is translating proteins.[33]
The limited window hypothesis offers a more general explanation for the retention of genes in non-photosynthetic plastids.[53] According to this hypothesis, genes are transferred to the nucleus following the disturbance of organelles.[34] Disturbance was common in the early stages of endosymbiosis, however, once the host cell gained control of organelle division, eukaryotes could evolve to have only one plastid per cell. Having only one plastid severely limits gene transfer[33] as the lysis of the single plastid would likely result in cell death.[33][53] Consistent with this hypothesis, organisms with multiple plastids show an 80-fold increase in plastid-to-nucleus gene transfer compared with organisms with single plastids.[53]
Evidence
There are many lines of evidence that mitochondria and plastids including chloroplasts arose from bacteria.[54][55][56][57][58]
- New mitochondria and plastids are formed only through binary fission, the form of cell division used by bacteria and archaea.[59]
- If a cell's mitochondria or chloroplasts are removed, the cell does not have the means to create new ones.[60] In some algae, such as Euglena, the plastids can be destroyed by certain chemicals or prolonged absence of light without otherwise affecting the cell: the plastids do not regenerate.
- Transport proteins called porins are found in the outer membranes of mitochondria and chloroplasts and are also found in bacterial cell membranes.[61][62][63]
- A membrane lipid cardiolipin is exclusively found in the inner mitochondrial membrane and bacterial cell membranes.[64]
- Some mitochondria and some plastids contain single circular DNA molecules that are similar to the DNA of bacteria both in size and structure.[65]
- Genome comparisons suggest a close relationship between mitochondria and Alphaproteobacteria.[66]
- Genome comparisons suggest a close relationship between plastids and cyanobacteria.[67]
- Many genes in the genomes of mitochondria and chloroplasts have been lost or transferred to the nucleus of the host cell. Consequently, the chromosomes of many eukaryotes contain genes that originated from the genomes of mitochondria and plastids.[65]
- Mitochondria and plastids contain their own ribosomes; these are more similar to those of bacteria (70S) than those of eukaryotes.[68]
- Proteins created by mitochondria and chloroplasts use N-formylmethionine as the initiating amino acid, as do proteins created by bacteria but not proteins created by eukaryotic nuclear genes or archaea.[69][70]
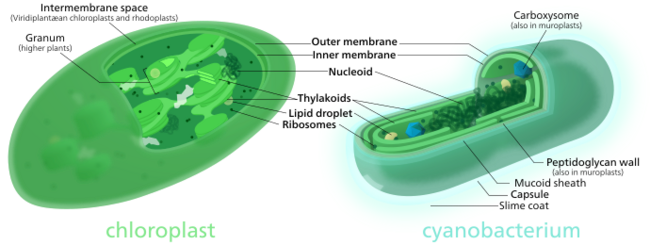
Secondary endosymbiosis
Primary endosymbiosis involves the engulfment of a cell by another free living organism. Secondary endosymbiosis occurs when the product of primary endosymbiosis is itself engulfed and retained by another free living eukaryote. Secondary endosymbiosis has occurred several times and has given rise to extremely diverse groups of algae and other eukaryotes. Some organisms can take opportunistic advantage of a similar process, where they engulf an alga and use the products of its photosynthesis, but once the prey item dies (or is lost) the host returns to a free living state. Obligate secondary endosymbionts become dependent on their organelles and are unable to survive in their absence. A secondary endosymbiosis event involving an ancestral red alga and a heterotrophic eukaryote resulted in the evolution and diversification of several other photosynthetic lineages including Cryptophyta, Haptophyta, Stramenopiles (or Heterokontophyta), and Alveolata.[71]
A possible secondary endosymbiosis has been observed in process in the heterotrophic protist Hatena. This organism behaves like a predator until it ingests a green alga, which loses its flagella and cytoskeleton but continues to live as a symbiont. Hatena meanwhile, now a host, switches to photosynthetic nutrition, gains the ability to move towards light, and loses its feeding apparatus.[72]
Despite the diversity of organisms containing plastids, the morphology, biochemistry, genomic organisation, and molecular phylogeny of plastid RNAs and proteins suggest a single origin of all extant plastids – although this theory is still debated.[73][74]
Some species including Pediculus humanus (lice) have multiple chromosomes in the mitochondrion. This and the phylogenetics of the genes encoded within the mitochondrion suggest that mitochondria have multiple ancestors, that these were acquired by endosymbiosis on several occasions rather than just once, and that there have been extensive mergers and rearrangements of genes on the several original mitochondrial chromosomes.[75]
Date
The question of when the transition from prokaryotic to eukaryotic form occurred and when the first crown group eukaryotes appeared on earth is still unresolved. The oldest known body fossils that can be positively assigned to the Eukaryota are acanthomorphic acritarchs from the 1.631 Gya Deonar Formation of India.[76] These fossils can still be identified as derived post-nuclear eukaryotes with a sophisticated, morphology-generating cytoskeleton sustained by mitochondria.[77] This fossil evidence indicates that endosymbiotic acquisition of alphaproteobacteria must have occurred before 1.6 Gya. Molecular clocks have also been used to estimate the last eukaryotic common ancestor, however these methods have large inherent uncertainty and give a wide range of dates. Reasonable results include the estimate of c. 1.8 Gya.[78] A 2.3 Gya estimate[79] also seems reasonable, and has the added attraction of coinciding with one of the most pronounced biogeochemical perturbations in Earth history, the early Palaeoproterozoic Great Oxygenation Event. The marked increase in atmospheric oxygen concentrations at that time has been suggested as a contributing cause of eukaryogenesis, inducing the evolution of oxygen-detoxifying mitochondria.[80] Alternatively, the Great Oxidation Event might be a consequence of eukaryogenesis, and its impact on the export and burial of organic carbon.[81]
See also
- Angomonas deanei, a protozoan that harbours an obligate bacterial symbiont
- Hatena arenicola, a species that appears to be in the process of acquiring an endosymbiont
- Hydrogen hypothesis, the theory that mitochondria were acquired by hydrogen-dependent archaea, their endosymbionts being facultatively anaerobic bacteria
- Kleptoplasty, the sequestering of plastids from ingested algae
- Mixotricha paradoxa, which itself is a symbiont, contains numerous endosymbiotic bacteria
- Parakaryon myojinensis, a possible result of endosymbiosis independent of eukaryotes
- Parasite Eve, fiction about endosymbiosis
- Strigomonas culicis, another protozoan that harbours an obligate bacterial symbiont
- Viral eukaryogenesis, hypothesis that the cell nucleus originated from endosymbiosis
References
- ↑ Latorre, A.; Durban, A.; Moya, A.; Pereto, J. (2011). "The role of symbiosis in eukaryotic evolution". Origins and Evolution of Life: An astrobiological perspective. Cambridge: Cambridge University Press. pp. 326–339. ISBN 978-0-521-76131-4. https://books.google.com/books?id=m3oFebknu1cC&pg=PA326. Retrieved 27 August 2017.
- ↑ "Serial endosymbiotic theory (SET)". http://flax.nzdl.org/greenstone3/flax;jsessionid=75E576A9FC9BB417BEC09D574631A027?a=d&c=BAWELS&d=D2065&dt=simple&p.a=b&p.s=ClassifierBrowse.
- ↑ 3.0 3.1 Cornish-Bowden, Athel (7 December 2017). "Lynn Margulis and the origin of the eukaryotes". Journal of Theoretical Biology. The origin of mitosing cells: 50th anniversary of a classic paper by Lynn Sagan (Margulis) 434: 1. doi:10.1016/j.jtbi.2017.09.027. PMID 28992902. Bibcode: 2017JThBi.434....1C. https://www.sciencedirect.com/science/article/pii/S0022519317304459.
- ↑ 4.0 4.1 "Mereschkowsky's Tree of Life". Scientific American. https://www.scientificamerican.com/article/mereschkowskys-tree-of-li/#. Retrieved 1 May 2017.
- ↑ Mereschkowski, Konstantin (15 September 1905). "Über Natur und Ursprung der Chromatophoren im Pflanzenreiche" (in de). Biologisches Centralblatt 25 (18): 593–604. https://babel.hathitrust.org/cgi/pt?id=uc1.31822009245838;view=1up;seq=603.
- ↑ See:
- Mereschkowski, Konstantin (15 April 1910). "Theorie der zwei Plasmaarten als Grundlage der Symbiogenesis, einer neuen Lehre von der Entstehung der Organismen" (in de). Biologisches Centralblatt 30 (8): 278–288. https://babel.hathitrust.org/cgi/pt?id=uc1.31822009245903;view=1up;seq=292.
- Mereschkowsky, Konstantin (1 May 1910). "Theorie der zwei Plasmaarten als Grundlage der Symbogenesis, einer neuen Lehre von der Entstehung der Organismen" (in de). Biologisches Centralblatt 30 (9): 289–303. https://babel.hathitrust.org/cgi/pt?id=uc1.31822009245903;view=1up;seq=303.
- Mereschkowski, Konstantin (15 May 1910). "Theorie der zwei Plasmaarten als Grundlage der Symbiogenesis, einer neuen Lehre von der Entstehung der Organismen" (in de). Biologisches Centralblatt 30 (10): 321–347. https://babel.hathitrust.org/cgi/pt?id=uc1.31822009245903;view=1up;seq=335.
- Mereschkowsky, Konstantin (1 June 1910). "Theorie der zwei Plasmaarten als Grundlage der Symbiogenesis, einer neuen Lehre von der Entstehung der Organismen" (in de). Biologisches Centralblatt 30 (11): 353–367. https://babel.hathitrust.org/cgi/pt?id=uc1.31822009245903;view=1up;seq=367.
- ↑ Martin, William F.; Roettger, Mayo; Kloesges, Thorsten et al.. "Modern endosymbiotic theory: Getting lateral gene transfer into the equation". Journal of Endocytobiosis and Cell Research 23: 1–5. http://www.molevol.hhu.de/fileadmin/redaktion/Fakultaeten/Mathematisch-Naturwissenschaftliche_Fakultaet/Biologie/Institute/Molekulare_Evolution/Dokumente/martin001.pdf. Retrieved 2015-07-20.(journal URL: [1] )
- ↑ See:
- Schimper, A. F. W. (16 February 1883). "Ueber die Entwicklung der Chlorophyllkörner und Farbkörper" (in de). Botanische Zeitung 41 (7): 105–114. https://babel.hathitrust.org/cgi/pt?id=hvd.32044106391576;view=1up;seq=91. From p. 105: "Inzwischen theilte mir Herr Professor Schmitz mit, dass … die höheren Pflanzen sich ebenso verhalten würden." (Meanwhile, Prof. Schmitz reported to me that among algae, the creation of chlorophyll granules from the cell plasm doesn't occur, but that they arise exclusively from one another by division. The spores receive from the mother plant chlorophyll granules, which create, by division, all of the chlorophyll granules of the plants that arises from them [i.e., the spores]. This finding in algae made it seem likely to Prof. Schmitz that the higher plants would behave likewise.) From p. 106: "Meine Untersuchungen haben ergeben, … aus dem Scheitelmeristem sich entwickelnden Gewebe erzeugen." (My investigations have revealed that the vegetation points [i.e., points of vegetative growth] always contain differentiated chlorophyll bodies or their colorless rudiments; that they arise not by creation from the cell plasm, but from one another by division, and that they create all chlorophyll bodies and starch-forming [bodies] of the tissues developing from the apical meristem.) From p. 112, footnote 2: "Sollte es sich definitiv bestätigen, … an eine Symbiose erinnern." (If it should definitely be confirmed that the plastids in egg cells are not formed anew, then their relation to the organism containing them would somewhat suggest a symbiosis.)
- Schimper, A. F. W. (23 February 1883). "Ueber die Entwicklung der Chlorophyllkörner und Farbkörper" (in de). Botanische Zeitung 41 (8): 121–131. https://babel.hathitrust.org/cgi/pt?id=hvd.32044106391576;view=1up;seq=99.
- Schimper, A. F. W. (2 March 1883). "Ueber die Entwicklung der Chlorophyllkörner und Farbkörper" (in de). Botanische Zeitung 41 (9): 137–146. https://babel.hathitrust.org/cgi/pt?id=hvd.32044106391576;view=1up;seq=107.
- Schimper, A. F. W. (9 March 1883). "Ueber die Entwicklung der Chlorophyllkörner und Farbkörper" (in de). Botanische Zeitung 41 (10): 153–162. https://babel.hathitrust.org/cgi/pt?id=hvd.32044106391576;view=1up;seq=115.
- ↑ Portier, Paul (1918) (in fr). Les Symbiotes. Paris, France: Masson et Cie.. p. 293. https://babel.hathitrust.org/cgi/pt?id=mdp.39015011399261;view=1up;seq=325. From p. 293: "Cette modification dans les rapports des appareils nucléaire et mitochondrial peut être le résultat de deux mécanismes. … Cette la parthénogénèse." (This modification in the relations of the nuclear and mitochondrial systems could be the result of two mechanisms: (a) There is a combination of two factors: contribution of new symbionts by the spermatozoid and reduction division. That is fertilization. (b) A single factor exists: reduction division: in this case, the egg contains sufficiently active symbionts. That is parthenogenesis.)
- ↑ Lane, Nick (2005). Power, Sex, Suicide. Mitochondria and the Meaning of Life. New York: Oxford University Press. p. 14. ISBN 9780199205646. https://archive.org/details/powersexsuicidem0000lane.
- ↑ Wallin, Ivan E. (1923). "The Mitochondria Problem". The American Naturalist 57 (650): 255–61. doi:10.1086/279919.
- ↑ Wallin, Ivan E. (1927). Symbionticism and the origin of species. Baltimore: Williams & Wilkins. pp. 117. https://archive.org/stream/symbionticismori00wall#page/116/mode/2up.
- ↑ Margulis, Lynn (2011). "Symbiogenesis. A new principle of evolution rediscovery of Boris Mikhaylovich Kozo-Polyansky (1890–1957)". Paleontological Journal 44 (12): 1525–1539. doi:10.1134/S0031030110120087. https://works.bepress.com/cgi/viewcontent.cgi?article=1019&context=lynn_margulis.
- ↑ Kozo-Polyansky, Boris Mikhaylovich (1924) (in ru). Новый принцип биологии. Очерк теории симбиогенеза. Moscow and Leningrad (St. Petersburg), Russia: Пучина (Puchina).
- English translation: Kozo-Polyansky, Boris Mikhaylovich (2010). Margulis, Lynn. ed. Symbiogenesis: A New Principle of Evolution. Cambridge, Massachusetts: Harvard University Press.
- Reviewed in: Niklas, Karl J. (2010). "Boris M. Kozo-Polyansky, Symbiogenesis: A New Principle of Evolution". Symbiosis 52 (1): 49–50. doi:10.1007/s13199-010-0098-7. Bibcode: 2010Symbi..52...49N.
- ↑ Corning, Peter A. (2010). Holistic Darwinism: Synergy, Cybernetics, and the Bioeconomics of Evolution. Chicago: University of Chicago Press. p. 81. ISBN 978-0-22611-633-4. https://books.google.com/books?id=NDeW4UNRmkwC.
- ↑ Ris, Hans; Plaut, Walter (June 1962). "Ultrastructure of DNA-containing areas in the chloroplast of Chlamydomonas". The Journal of Cell Biology 13 (3): 383–91. doi:10.1083/jcb.13.3.383. PMID 14492436.
- ↑ Ris, Hans; Singh, R. N. (January 1961). "Electron microscope studies on blue-green algae". The Journal of Biophysical and Biochemical Cytology 9 (1): 63–80. doi:10.1083/jcb.9.1.63. PMID 13741827.
- ↑ Stocking, C.; Gifford, E. (1959). "Incorporation of thymidine into chloroplasts of Spirogyra". Biochem. Biophys. Res. Commun. 1 (3): 159–64. doi:10.1016/0006-291X(59)90010-5.
- ↑ Sagan, Lynn (March 1967). "On the origin of mitosing cells". Journal of Theoretical Biology 14 (3): 255–74. doi:10.1016/0022-5193(67)90079-3. PMID 11541392. Bibcode: 1967JThBi..14..225S.
- ↑ Margulis, Lynn; Sagan, Dorion (1997). Microcosmos: Four Billion Years of Microbial Evolution. Berkeley, Los Angeles, London: University of California Press. p. 29. ISBN 0-520-21064-6. https://books.google.com/books?id=eo_sMMRxgAUC.
- ↑ Gabaldón, Toni; Snel, Berend; Zimmeren, Frank van et al. (23 March 2006). "Origin and evolution of the peroxisomal proteome". Biology Direct 1 (1): 8. doi:10.1186/1745-6150-1-8. PMID 16556314. (Provides evidence that contradicts an endosymbiotic origin of peroxisomes, and suggests instead that they originate evolutionarily from the endoplasmic reticulum)
- ↑ "Supertrees disentangle the chimerical origin of eukaryotic genomes". Molecular Biology and Evolution 24 (8): 1752–1760. August 2007. doi:10.1093/molbev/msm095. PMID 17504772.
- ↑ 23.0 23.1 Keeling, P. J.; Archibald, J. M. (April 2008). "Organelle evolution: what's in a name?". Current Biology 18 (8): R345-7. doi:10.1016/j.cub.2008.02.065. PMID 18430636.
- ↑ Syvanen, Michael; Kado, Clarence I. (30 January 2002). Horizontal Gene Transfer. Academic Press. p. 405. ISBN 978-0126801262.
- ↑ 25.00 25.01 25.02 25.03 25.04 25.05 25.06 25.07 25.08 25.09 25.10 25.11 25.12 25.13 25.14 25.15 Timmis, Jeremy N.; Ayliffe, Michael A.; Huang, Chun Y.; Martin, William (2004). "Endosymbiotic gene transfer: organelle genomes forge eukaryotic chromosomes". Nature Reviews Genetics 5 (2): 123–135. doi:10.1038/nrg1271. PMID 14735123.
- ↑ "Mitochondria Share an Ancestor With SAR11, a Globally Significant Marine Microbe". July 25, 2011. https://www.sciencedaily.com/releases/2011/07/110725190046.htm.
- ↑ Thrash, J. Cameron; Boyd, Alex; Huggett, Megan J. et al. (14 June 2011). "Phylogenomic evidence for a common ancestor of mitochondria and the SAR11 clade". Scientific Reports 1 (1): 13. doi:10.1038/srep00013. PMID 22355532. Bibcode: 2011NatSR...1E..13T.
- ↑ Deusch, O.; Landan, G.; Roettger, M. et al. (14 February 2008). "Genes of Cyanobacterial Origin in Plant Nuclear Genomes Point to a Heterocyst-Forming Plastid Ancestor". Molecular Biology and Evolution 25 (4): 748–761. doi:10.1093/molbev/msn022. PMID 18222943.
- ↑ Ochoa de Alda, Jesús A. G.; Esteban, Rocío; Diago, María Luz et al. (15 September 2014). "The plastid ancestor originated among one of the major cyanobacterial lineages". Nature Communications 5 (1): 4937. doi:10.1038/ncomms5937. PMID 25222494. Bibcode: 2014NatCo...5.4937O.
- ↑ 30.0 30.1 30.2 Lila Koumandou, V.; Nisbet, R. Ellen R.; Barbrook, Adrian C. et al. (May 2004). "Dinoflagellate chloroplasts—where have all the genes gone?". Trends in Genetics 20 (5): 261–267. doi:10.1016/j.tig.2004.03.008. PMID 15109781.
- ↑ Taanman, J. W. (February 1999). "The mitochondrial genome: structure, transcription, translation and replication". Biochimica et Biophysica Acta (BBA) - Bioenergetics 1410 (2): 103–23. doi:10.1016/S0005-2728(98)00161-3. PMID 10076021.
- ↑ Nowack, E. C.; Melkonian, M.; Glockner, G. (March 2008). "Chromatophore genome sequence of Paulinella sheds light on acquisition of photosynthesis by eukaryotes". Current Biology 18 (6): 410–8. doi:10.1016/j.cub.2008.02.051. PMID 18356055.
- ↑ 33.00 33.01 33.02 33.03 33.04 33.05 33.06 33.07 33.08 33.09 Barbrook, Adrian C.; Howe, Christopher J.; Purton, Saul (February 2006). "Why are plastid genomes retained in non-photosynthetic organisms?". Trends in Plant Science 11 (2): 101–8. doi:10.1016/j.tplants.2005.12.004. PMID 16406301.
- ↑ 34.0 34.1 34.2 34.3 34.4 34.5 34.6 Leister, D. (December 2005). "Origin, evolution and genetic effects of nuclear insertions of organelle DNA". Trends in Genetics 21 (12): 655–63. doi:10.1016/j.tig.2005.09.004. PMID 16216380. http://edoc.mpg.de/277780.
- ↑ Keeling, P. J. (October 2004). "Diversity and evolutionary history of plastids and their hosts". American Journal of Botany 91 (10): 1481–93. doi:10.3732/ajb.91.10.1481. PMID 21652304.
- ↑ 36.0 36.1 36.2 Archibald, J. M. (January 2009). "The puzzle of plastid evolution". Current Biology 19 (2): R81–R88. doi:10.1016/j.cub.2008.11.067. PMID 19174147.
- ↑ 37.0 37.1 37.2 Ford Doolittle, W (1998-12-01). "You are what you eat: a gene transfer ratchet could account for bacterial genes in eukaryotic nuclear genomes". Trends in Genetics 14 (8): 307–311. doi:10.1016/S0168-9525(98)01494-2. ISSN 0168-9525. PMID 9724962. https://www.sciencedirect.com/science/article/pii/S0168952598014942.
- ↑ Zimorski, Verena; Ku, Chuan; Martin, William F; Gould, Sven B (2014). "Endosymbiotic theory for organelle origins". Current Opinion in Microbiology 22: 38–48. doi:10.1016/j.mib.2014.09.008. PMID 25306530.
- ↑ "Mitochondria, Cell Energy, ATP Synthase: Learn Science at Scitable". https://www.nature.com/scitable/topicpage/mitochondria-14053590.
- ↑ 40.0 40.1 Gruber, A. (January 2019). "What's in a name? How organelles of endosymbiotic origin can be distinguished from endosymbionts". Microbial Cell 6 (2): 123–133. doi:10.15698/mic2019.02.668. PMID 30740457.
- ↑ 41.0 41.1 41.2 41.3 41.4 41.5 Gould, Sven B.; Garg, Sriram G.; Martin, William F. (July 2016). "Bacterial Vesicle Secretion and the Evolutionary Origin of the Eukaryotic Endomembrane System". Trends in Microbiology 24 (7): 525–534. doi:10.1016/j.tim.2016.03.005. PMID 27040918. https://zenodo.org/record/3450278.[yes|permanent dead link|dead link}}]
- ↑ 42.0 42.1 Martin, William F.; Garg, Sriram; Zimorski, Verena (September 2015). "Endosymbiotic theories for eukaryote origin". Philosophical Transactions of the Royal Society of London. Series B, Biological Sciences 370 (1678): 20140330. doi:10.1098/rstb.2014.0330. PMID 26323761.
- ↑ Garavís, Miguel; González, Carlos; Villasante, Alfredo (June 2013). "On the origin of the eukaryotic chromosome: the role of noncanonical DNA structures in telomere evolution". Genome Biology and Evolution 5 (6): 1142–50. doi:10.1093/gbe/evt079. PMID 23699225.
- ↑ "Typical prokaryotic (left) and eukaryotic (right) cells: Learn Science at Scitable". https://www.nature.com/scitable/content/typical-prokaryotic-left-and-eukaryotic-right-cells-14665031.
- ↑ Devos, Damien P.; Gräf, Ralph; Field, Mark C. (June 2014). "Evolution of the nucleus". Current Opinion in Cell Biology 28 (100): 8–15. doi:10.1016/j.ceb.2014.01.004. PMID 24508984.
- ↑ Wilson, Katherine L.; Dawson, Scott C. (October 2011). "Evolution: functional evolution of nuclear structure". Journal of Cell Biology 195 (2): 171–81. doi:10.1083/jcb.201103171. PMID 22006947.
- ↑ Bernstein, H.; Bernstein, C. (2017). "Sexual communication in archaea, the precursor to meiosis". in Witzany, G.. Biocommunication of Archaea. Springer International Publishing. pp. 103–117. doi:10.1007/978-3-319-65536-9. ISBN 978-3-319-65535-2.
- ↑ Gabaldón, T.; Huynen, M. A. (August 2003). "Reconstruction of the proto-mitochondrial metabolism". Science 301 (5633): 609. doi:10.1126/science.1085463. PMID 12893934.
- ↑ Liashkovich, Ivan; Shahin, Victor (August 2017). "Functional implication of the common evolutionary origin of nuclear pore complex and endomembrane management systems". Seminars in Cell and Developmental Biology 68: 10–17. doi:10.1016/j.semcdb.2017.04.006. PMID 28473267.
- ↑ 50.0 50.1 Howe, Christopher J. (May 2008). "Cellular evolution: what's in a mitochondrion?". Current Biology 18 (10): R429–R431. doi:10.1016/j.cub.2008.04.007. PMID 18492476.
- ↑ López-García, Purificación; Moreira, David (2020-04-27). "The Syntrophy hypothesis for the origin of eukaryotes revisited". Nature Microbiology 5 (5): 655–667. doi:10.1038/s41564-020-0710-4. ISSN 2058-5276. PMID 32341569. https://hal.archives-ouvertes.fr/hal-02988531/file/Final_submitted_ms.pdf.
- ↑ Giannakis, Konstantinos; Arrowsmith, Samuel J.; Richards, Luke et al. (16 September 2022). "Evolutionary inference across eukaryotes identifies universal features shaping organelle gene retention". Cell Systems 13 (11): 874–884.e5. doi:10.1016/j.cels.2022.08.007. PMID 36115336.
- ↑ 53.0 53.1 53.2 53.3 Lane, Nick (2011). "Plastids, genomes, and the probability of gene transfer". Genome Biology and Evolution 3: 372–374. doi:10.1093/gbe/evr003. PMID 21292628.
- ↑ Kimball, J. 2010. Kimball's Biology Pages. Accessed October 13, 2010. An online open source biology text by Harvard professor, and author of a general biology text, John W. Kimball.
- ↑ Reece, J., Lisa A. Urry, Michael L. Cain, Steven A. Wasserman, Peter V. Minorsky, Robert B. Jackson, 2010. Campbell Biology. 9th Edition Benjamin Cummings; 9th Ed. (October 7, 2010)
- ↑ Raven, P.; Johnson, George; Mason, Kenneth et al. (January 14, 2010). Biology (9th ed.). McGraw-Hill.
- ↑ Gray, M. W. (1992). "The endosymbiont hypothesis revisited". International Review of Cytology 141: 233–357. doi:10.1016/S0074-7696(08)62068-9. ISBN 9780123645449. PMID 1452433.
- ↑ Zimorski, V.; Ku, C.; Martin, W. F.; Gould, S. B. (December 2014). "Endosymbiotic theory for organelle origins". Current Opinion in Microbiology 22: 38–48. doi:10.1016/j.mib.2014.09.008. PMID 25306530.
- ↑ Margolin, William (November 2005). "FtsZ and the division of prokaryotic cells and organelles". Nature Reviews. Molecular Cell Biology 6 (11): 862–71. doi:10.1038/nrm1745. PMID 16227976.
- ↑ Wise, Robert R.; Hoober, J. Kenneth (2007). Structure and function of plastids. Berlin: Springer. p. 104. ISBN 9781402065705. https://books.google.com/books?id=FKeCVPbJ3asC&pg=PA104.
- ↑ Fischer, K.; Weber, A.; Brink, S. et al. (October 1994). "Porins from plants. Molecular cloning and functional characterization of two new members of the porin family". The Journal of Biological Chemistry 269 (41): 25754–60. doi:10.1016/S0021-9258(18)47312-7. PMID 7523392.
- ↑ Zeth, K.; Thein, M. (October 2010). "Porins in prokaryotes and eukaryotes: common themes and variations". The Biochemical Journal 431 (1): 13–22. doi:10.1042/BJ20100371. PMID 20836765.
- ↑ Fairman, J. W.; Noinaj, N.; Buchanan, S. K. (August 2011). "The structural biology of β-barrel membrane proteins: a summary of recent reports". Current Opinion in Structural Biology 21 (4): 523–331. doi:10.1016/j.sbi.2011.05.005. PMID 21719274.
- ↑ Mileykovskaya, E.; Dowhan, W. (October 2009). "Cardiolipin membrane domains in prokaryotes and eukaryotes". Biochimica et Biophysica Acta (BBA) - Biomembranes 1788 (10): 2084–91. doi:10.1016/j.bbamem.2009.04.003. PMID 19371718.
- ↑ 65.0 65.1 Timmis, Jeremy; Ayliffe, Michael; Huang, Chun; Martin, William (February 2004). "Endosymbiotic gene transfer: organelle genomes forge eukaryotic chromosomes". Nature Reviews. Genetics 5 (2): 123–35. doi:10.1038/nrg1271. PMID 14735123.
- ↑ Munoz-Gomez, Sergio; Susko, Edward; Williamson, Kelsey et al. (January 2022). "Site-and-branch-heterogeneous analyses of an expanded dataset favour mitochondria as sister to known Alphaproteobacteria". Nature Ecology and Evolution 6 (3): 253–62. doi:10.1038/s41559-021-01638-2. PMID 35027725. Bibcode: 2022NatEE...6..253M.
- ↑ Dagan, Tal; Roettger, Mayo; Stucken, Karina et al. (2013). "Genomes of Stigonematalean cyanobacteria (subsection V) and the evolution of oxygenic photosynthesis from prokaryotes to plastids". Genome Biology and Evolution 5 (1): 31–44. doi:10.1093/gbe/evs117. PMID 23221676.
- ↑ Manuell, Andrea L.; Quispe, Joel; Mayfield, Stephen P. (August 2007). "Structure of the chloroplast ribosome: novel domains for translation regulation". PLOS Biology 5 (8): e209. doi:10.1371/journal.pbio.0050209. PMID 17683199.
- ↑ Schwartz, James H.; Meyer, Ralph; Eisenstadt, Jerome M.; Brawerman, George (May 1967). "Involvement of N-formylmethionine in initiation of protein synthesis in cell-free extracts of Euglena gracilis". Journal of Molecular Biology 25 (3): 571–4. doi:10.1016/0022-2836(67)90210-0. PMID 5340700.
- ↑ Smith, A.E.; Marcker, K.A. (December 1968). "N-formylmethionyl transfer RNA in mitochondria from yeast and rat liver". Journal of Molecular Biology 38 (2): 241–3. doi:10.1016/0022-2836(68)90409-9. PMID 5760639.
- ↑ McFadden, G. I. (2001). "Primary and secondary endosymbiosis and the origin of plastids". Journal of Phycology 37 (6): 951–959. doi:10.1046/j.1529-8817.2001.01126.x. Bibcode: 2001JPcgy..37..951M.
- ↑ Okamoto, N.; Inouye, I. (October 2005). "A secondary symbiosis in progress?". Science 310 (5746): 287. doi:10.1126/science.1116125. PMID 16224014. https://www.researchgate.net/publication/7542363.
- ↑ McFadden, G. I.; van Dooren, G. G. (July 2004). "Evolution: red algal genome affirms a common origin of all plastids". Current Biology 14 (13): R514-6. doi:10.1016/j.cub.2004.06.041. PMID 15242632.
- ↑ Gould, Sven B.; Waller, Ross F.; McFadden, Geoffrey I. (2008). "Plastid evolution". Annual Review of Plant Biology 59 (1): 491–517. doi:10.1146/annurev.arplant.59.032607.092915. PMID 18315522.
- ↑ Georgiades, K.; Raoult, D. (October 2011). "The rhizome of Reclinomonas americana, Homo sapiens, Pediculus humanus and Saccharomyces cerevisiae mitochondria". Biology Direct 6: 55. doi:10.1186/1745-6150-6-55. PMID 22014084.
- ↑ Prasad, Pijai (August 2005). "Organic-walled microfossils from the Proterozoic Vindhyan Supergroup of Son Valley, Madhya Pradesh, India". Paleobotanist 54. http://14.139.63.228:8080/pbrep/bitstream/123456789/1084/1/PbV54_13.pdf.
- ↑ Butterfield, Nicholas J. (2014-11-26). "Early evolution of the Eukaryota". Palaeontology 58 (1): 5–17. doi:10.1111/pala.12139.
- ↑ Parfrey, Laura Wegener; Lahr, Daniel J. G.; Knoll, Andrew H.; Katz, Laura A. (August 2011). "Estimating the timing of early eukaryotic diversification with multigene molecular clocks". Proceedings of the National Academy of Sciences of the United States of America 108 (33): 13624–9. doi:10.1073/pnas.1110633108. PMID 21810989. Bibcode: 2011PNAS..10813624P.
- ↑ Hedges, S. Blair; Blair, Jaime E.; Venturi, Maria L.; Shoe, Jason L. (January 2004). "A molecular timescale of eukaryote evolution and the rise of complex multicellular life". BMC Evolutionary Biology 4: 2. doi:10.1186/1471-2148-4-2. PMID 15005799.
- ↑ Gross, Jeferson; Bhattacharya, Debashish (August 2010). "Uniting sex and eukaryote origins in an emerging oxygenic world". Biology Direct 5: 53. doi:10.1186/1745-6150-5-53. PMID 20731852.
- ↑ Butterfield, Nicholas J. (1997). "Plankton ecology and the Proterozoic-Phanerozoic transition". Paleobiology 23 (2): 247–262. doi:10.1017/S009483730001681X. Bibcode: 1997Pbio...23..247B.
Further reading
- Alberts, Bruce (2002). Molecular Biology of the Cell. New York: Garland Science. ISBN 978-0-8153-3218-3. (General textbook)
- Brinkman, Fiona S. L.; Blanchard, Jeffrey L.; Cherkasov, Artem et al. (August 2002). "Evidence that plant-like genes in Chlamydia species reflect an ancestral relationship between Chlamydiaceae, cyanobacteria, and the chloroplast". Genome Research 12 (8): 1159–67. doi:10.1101/gr.341802. PMID 12176923.
- Cohen, W. E.; Gardner, R. S. (1959). "Viral Theory and Endosymbiosis". http://www.psychoneuroendocrinology.com/symbiosis.pdf. (Discusses theory of origin of eukaryotic cells by incorporating mitochondria and chloroplasts into anaerobic cells with emphasis on 'phage bacterial and putative viral mitochondrial/chloroplast interactions.)
- Jarvis, P. (April 2001). "Intracellular signalling: the chloroplast talks!". Current Biology 11 (8): R307-10. doi:10.1016/S0960-9822(01)00171-3. PMID 11369220. (Recounts evidence that chloroplast-encoded proteins affect transcription of nuclear genes, as opposed to the more well-documented cases of nuclear-encoded proteins that affect mitochondria or chloroplasts.)
- Blanchard, J. L.; Lynch, M. (July 2000). "Organellar genes: why do they end up in the nucleus?". Trends in Genetics 16 (7): 315–20. doi:10.1016/S0168-9525(00)02053-9. PMID 10858662. (Discusses theories on how mitochondria and chloroplast genes are transferred into the nucleus, and also what steps a gene needs to go through in order to complete this process.)
- Okamoto, N.; Inouye, I. (October 2005). "A secondary symbiosis in progress?". Science 310 (5746): 287. doi:10.1126/science.1116125. PMID 16224014.
- Understanding Science Team. "Cells within cells: An extraordinary claim with extraordinary evidence". University of California, Berkeley. http://undsci.berkeley.edu/lessons/pdfs/endosymbiosis.pdf.
External links
 |

