Biology:Sucrose-phosphate synthase
| sucrose-phosphate synthase | |||||||||
|---|---|---|---|---|---|---|---|---|---|
 Sucrose phosphate synthase monomer, Halothermothrix orenii | |||||||||
| Identifiers | |||||||||
| EC number | 2.4.1.14 | ||||||||
| CAS number | 9030-06-2 | ||||||||
| Databases | |||||||||
| IntEnz | IntEnz view | ||||||||
| BRENDA | BRENDA entry | ||||||||
| ExPASy | NiceZyme view | ||||||||
| KEGG | KEGG entry | ||||||||
| MetaCyc | metabolic pathway | ||||||||
| PRIAM | profile | ||||||||
| PDB structures | RCSB PDB PDBe PDBsum | ||||||||
| Gene Ontology | AmiGO / QuickGO | ||||||||
| |||||||||
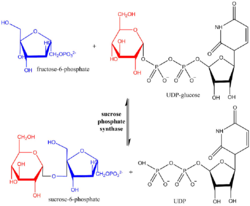
Sucrose-phosphate synthase (SPS) is a plant enzyme involved in sucrose biosynthesis. Specifically, this enzyme catalyzes the transfer of a hexosyl group from uridine diphosphate glucose (UDP-glucose) to D-fructose 6-phosphate to form UDP and D-sucrose-6-phosphate.[1][2] This reversible step acts as the key regulatory control point in sucrose biosynthesis, and is an excellent example of various key enzyme regulation strategies such as allosteric control and reversible phosphorylation.[3]
This enzyme participates in starch and sucrose metabolism.[2]
Nomenclature
This enzyme belongs to the family of glycosyltransferases, specifically the hexosyltransferases. The systematic name of this enzyme class is UDP-glucose:D-fructose 6-phosphate 2-alpha-D-glucosyltransferase. Other names in common use include UDP-glucose-fructose-phosphate glucosyltransferase, sucrosephosphate-UDP glucosyltransferase, UDP-glucose-fructose-phosphate glucosyltransferase, SPS, uridine diphosphoglucose-fructose phosphate glucosyltransferase, sucrose 6-phosphate synthase, sucrose phosphate synthetase, and sucrose phosphate-uridine diphosphate glucosyltransferase.
Structure
X-ray diffraction studies have revealed that the structure of Halothermothrix orenii SPS belongs to the GT-B fold family.[1] Like other GT-B proteins, SPS contains two Rossmann fold domains that are named the A domain and the B domain.[4] Generally, the structure of these domains are somewhat similar, as both contain central beta sheets that are surrounded by alpha helices. However, the A domain consists of eight parallel beta strands and seven alpha helices while the B domain contains six parallel beta strands and nine alpha helices. These domains are joined by residue loops to form a substrate binding cleft, where the glucosyl group acceptor binds.[1]
Although H. orenii is a non-photosynthetic bacterium, various studies indicate that the structure of its SPS is similar to plant SPS. First, antibodies with high specificities for plant SPS also target the bacterial SPS, indicating the structure is conserved enough for the antibody to recognize the enzyme as an antigen. Furthermore, genomic studies reveal that closely related plant homologues exhibit up to 54% sequence identities.[1]
Mechanism
In the open conformation of H. orenii SPS, fructose 6-phosphate forms hydrogen bonds with Gly-33 and Gln-35 residues in the A domain while UDP-glucose interacts with the B-domain. Crystal structures studies reveal that after binding, the two domains twist to narrow the entrance of the substrate binding cleft from 20 Å to 6 Å. In this closed conformation, the Gly-34 residue of the A domain interacts with UDP-glucose and forces the substrate to adapt a folded structure, facilitating its donation of the hexosyl group.[4]
After binding, fructose 6-phosphate will interact with UDP via a hydrogen bond, which lowers the activation energy of the reaction and stabilizes the transition state. Finally, the C1 atom of UDP-glucose undergoes nucleophilic attack by an oxygen atom in fructose 6-phosphate, resulting in glucosyl group transfer to fructose 6-phosphate. Whether or not this mechanism requires a divalent ion is currently unclear, but failed attempts to trap and detect the presence of the magnesium cation suggest that this mechanism is metal ion independent.[1][4]
Regulatory strategies
Phosphorylation
SPS-kinase reversibly phosphorylates a serine residue and subsequently deactivates SPS, In spinach and maize, the site of phosphorylation regulation has been identified as Ser158 and Ser162 respectively. While it is currently unclear if this seryl residue homolog in other plant SPSes is phosphorylated to suppress SPS activity, conservation of the neighboring residues has been observed in other plant species. This conserved sequence may potentially aid in recognition of a regulatory SPS-kinase. Once phosphorylated, the inactivated enzyme can be dephosphorylated and reactivated by SPS-phosphatase. Aside from controlling the levels of sucrose in the cell, regulation via phosphorylation can help the cell adapt to hyperosmotic conditions; in times of osmotic stress, the seryl residue is phosphorylated and enzyme activity decreases.[3] This regulation strategy also controls carbon flux from photosynthesis, as studies indicate the signal transduction pathway responsible for SPS activation responds to light stimulus.[5][6][7]
Allostery
Glucose 6-phosphate binds to an allosteric site, resulting in conformational changes to SPS that increase the enzyme's affinity for the glucosyl accepting substrate. Inorganic phosphate can also bind to this allosteric site, preventing glucose 6-phosphate activation of SPS. Like regulation via phosphorylation, this regulation strategy is also closely related to photosynthesis, as high rates of photosynthesis will deplete levels of inorganic phosphate and increase concentrations of glucose 6-phosphate in the chloroplast.[8] Overall, increased rates of photosynthesis will increase SPS activity.
Function
SPS plays a major role in partitioning carbon between sucrose and starch in photosynthetic and non-photosynthetic tissues, affecting the growth and development of the plant.[2][8][9] In ripening fruits, SPS is responsible for converting starch to sucrose and other soluble sugars.[10][11][12] Additionally, SPS is also active in cells that mostly degrade sucrose, participating in futile cycles that allow for large, rapid changes in sucrose flux.[3]
At low temperature, SPS activity and sucrose biosynthesis rates are increased. Sucrose accumulation is advantageous at low temperature, as sucrose is a form of energy storage that can be rapidly metabolized for respiratory purposes. Furthermore, increased amounts of sucrose can help the plant withstand freezing.[13]
References
- ↑ 1.0 1.1 1.2 1.3 1.4 "The structure of sucrose phosphate synthase from Halothermothrix orenii reveals its mechanism of action and binding mode". The Plant Cell 20 (4): 1059–72. April 2008. doi:10.1105/tpc.107.051193. PMID 18424616.
- ↑ 2.0 2.1 2.2 Levine, Martin (2011). Topics in Dental Biochemistry. Springer. pp. 17–27. ISBN 978-3-540-88115-5. https://archive.org/details/topicsdentalbioc00levi.
- ↑ 3.0 3.1 3.2 "ROLE AND REGULATION OF SUCROSE-PHOSPHATE SYNTHASE IN HIGHER PLANTS". Annual Review of Plant Physiology and Plant Molecular Biology 47: 431–444. June 1996. doi:10.1146/annurev.arplant.47.1.431. PMID 15012296.
- ↑ 4.0 4.1 4.2 "Structures and mechanisms of glycosyltransferases". Glycobiology 16 (2): 29R–37R. February 2006. doi:10.1093/glycob/cwj016. PMID 16037492.
- ↑ "Role of sucrose-phosphate synthase in sucrose metabolism in leaves". Plant Physiology 99 (4): 1275–8. August 1992. doi:10.1104/pp.99.4.1275. PMID 16669032.
- ↑ "Spinach Leaf Sucrose-Phosphate Synthase and Nitrate Reductase Are Phosphorylated/Inactivated by Multiple Protein Kinases in Vitro". Plant Physiology 108 (3): 1077–1082. July 1995. doi:10.1104/pp.108.3.1077. PMID 12228528.
- ↑ "Effects of Elevated Sucrose-Phosphate Synthase Activity on Photosynthesis, Assimilate Partitioning, and Growth in Tomato (Lycopersicon esculentum var UC82B)". Plant Physiology 101 (2): 535–543. February 1993. doi:10.1104/pp.101.2.535. PMID 12231708.
- ↑ 8.0 8.1 "Regulation of Spinach Leaf Sucrose Phosphate Synthase by Glucose-6-Phosphate, Inorganic Phosphate, and pH". Plant Physiology 73 (4): 989–94. December 1983. doi:10.1104/pp.73.4.989. PMID 16663357.
- ↑ "Role of sucrose-phosphate synthase in partitioning of carbon in leaves". Plant Physiology 71 (4): 818–21. April 1983. doi:10.1104/pp.71.4.818. PMID 16662913.
- ↑ Hubbard, Natalie L.; Pharr, D. Mason; Huber, Steven C. (1991). "Sucrose phosphate synthase and other sucrose etabolizing enzymes in fruits of various species". Physiologia Plantarum 82 (2): 191–196. doi:10.1111/j.1399-3054.1991.tb00080.x.
- ↑ "Sucrose Phosphate Synthase and Acid Invertase as Determinants of Sucrose Concentration in Developing Muskmelon (Cucumis melo L.) Fruits". Plant Physiology 91 (4): 1527–34. December 1989. doi:10.1104/pp.91.4.1527. PMID 16667212.
- ↑ "Sucrose Phosphate Synthase, Sucrose Synthase, and Invertase Activities in Developing Fruit of Lycopersicon esculentum Mill. and the Sucrose Accumulating Lycopersicon hirsutum Humb. and Bonpl". Plant Physiology 95 (2): 623–7. February 1991. doi:10.1104/pp.95.2.623. PMID 16668028.
- ↑ "Sucrose phosphate synthase and sucrose accumulation at low temperature". Plant Physiology 100 (1): 502–8. September 1992. doi:10.1104/pp.100.1.502. PMID 16652990.
 |