Medicine:Elastix (image registration)
| Developer(s) | Stefan Klein and Marius Staring et al. |
|---|---|
| Initial release | 2003 |
| Repository | github |
| Written in | C++ |
| Operating system | Linux, Windows, macOS |
| License | versions of elastix from 4.8 are under Apache 2.0, versions of elastix prior to 4.8 are under BSD license |
| Website | elastix |
Elastix is an image registration toolbox built upon the Insight Segmentation and Registration Toolkit (ITK).[1] It is entirely open-source and provides a wide range of algorithms employed in image registration problems. Its components are designed to be modular to ease a fast and reliable creation of various registration pipelines tailored for case-specific applications. It was first developed by Stefan Klein and Marius Staring under the supervision of Josien P.W. Pluim at Image Sciences Institute (ISI).[2] Its first version was command-line based, allowing the final user to employ scripts to automatically process big data-sets and deploy multiple registration pipelines with few lines of code. Nowadays, to further widen its audience, a version called SimpleElastix is also available,[3] developed by Kasper Marstal, which allows the integration of elastix with high level languages, such as Python, Java, and R.[4]
Image registration fundamentals
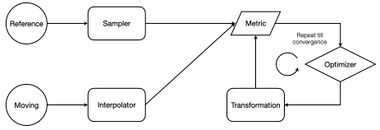
Image registration is a well-known technique in digital image processing that searches for the geometric transformation that, applied to a moving image, obtains a one-to-one map with a target image. Generally, the images acquired from different sensors (multimodal), time instants (multitemporal), and points of view (multiview) should be correctly aligned to proceed with further processing and feature extraction.[5] Even though there are a plethora of different approaches to image registration, the majority is composed of the same macro building blocks, namely the transformation, the interpolator, the metric, and the optimizer.[6] Registering two or more images can be framed as an optimization problem that requires multiple iterations to converge to the best solution. Starting from an initial transformation computed from the image moments the optimization process searches for the best transformation parameters based on the value of the selected similarity metric. The figure on the right shows the high-level representation of the registration of two images, where the reference remains constant during the entire process, while the moving one will be transformed according to the transformation parameters. In other words, the registration ends when the similarity metric, which is a mathematical function with a certain number of parameters to be optimized, reaches the optimal value which is highly dependent on the specific application.[6]
Main building blocks
Following the structure of the image registration workflow, the elastix toolbox proposes a modular solution that implements for each of the building blocks different algorithms, highly employed in medical image registration, and helps the final users to build their specific pipeline by selecting the most suitable algorithm for each of the main building blocks. Each block is easily configurable both by selecting pre-defined initialization values or by trying multiple sets of parameters and then choosing the most performing one. The registration is performed on images, and the elastix toolbox supports all the data formats supported by ITK, ranging from JPEG and PNG to medical standard formats such as DICOM and NIFTI. It also stores physical pixel spacing, the origin and the relative position to an external world reference system, when provided in the metadata, to facilitate the registration process, especially in medical field applications.[7]
Transformation
The transformation is an essential building block, since it defines the allowable transformations. In image registration, the main distinction can be done between parallel-to-parallel and parallel-to-non parallel (deformable) line mapping transformations. In the elastix toolbox, the final users can select one transformation or compose more transformations either through addition or via composition. Below are reported the different transformation models in order of increasing flexibility, along with the corresponding elastix class names between brackets.[7]
- Translation (TranslationTransform) allows only translations
- Rigid (EulerTransform) expands the translation adding rotations and the object is seen as a rigid body
- Similarity (SimilarityTransform) expands the rigid transformation by introducing isotropic scaling
- Affine (AffineTransform) expands the rigid transformation allowing both scaling and shear
- B-splines (BSplineTransform) is a deformable transformation usually preceded by a rigid or affine one[8]
- Thin-plate splines (SplineKernelTransform) is a deformable transformation belonging to the class of kernel-based transformations[9] that is a composition of and affine and a non-rigid part
Metric
The similarity metric is the mathematical function whose parameters should be optimized to reach the desired registration, and, during the process, it is computed multiple times. Below are reported the available metrics computed employing the reference and the transformed images and the corresponding elastix class names between brackets.[7]
- Mean squared difference (AdvancedMeanSquares) to be used for mono-modal applications
- Normalized correlation coefficient (AdvancedNormalizedCorrelation) to be used for images that have an intensity linear relationship
- Mutual information (AdvancedMattesMutualInformation) to be used for both mono- and multi-modal applications and optimized to reach better performance compared to the normalized version
- Normalized mutual information (NormalizedMutualInformation) for both mono- and multi-modal applications
- Kappa statistic (AdvancedKappaStatistic) to be used only for binary images
Sampler
For the computation of the similarity metrics, it is not always necessary to consider all the voxels and, sometimes, it can be useful to use only a fraction of the voxels of the images, i.e. to reduce the execution time for big input images.[10] Below are reported the available criteria for selecting a fraction of the voxels for the similarity metric computation and the corresponding elastix class names between brackets.[7]
- Full (Full) to employ all the voxels
- Grid (Grid) to employ a regular grid defined by the user to downsample the image
- Random (Random) to randomly select a percentage of voxels defined by the users (all voxels have equal probability to be selected)
- Random coordinate (RandomCoordinate) like the random criterion, but in this case also off-grid positions can be selected to simplify the optimization process
Interpolator
After the application of the transformation, it may occur that the voxels used for the similarity metric computation are at non-voxel positions, so intensity interpolation should be performed to ensure the correctness of the computed values. Below are reported the implemented interpolators and the corresponding elastix class names between brackets.[7]
- Nearest neighbor (NearestNeighborInterpolator) exploits little resources, but gives low quality results
- Linear (LinearInterpolator) is sufficient in general applications
- N-th order B-spline (BSplineInterpolator) can be used to increase the order N, increasing quality and computation time. N=0 and N=1 indicate the nearest neighbor and linear cases respectively.[11]
Optimizer
The optimizer defines the strategy employed for searching the best transformation parameter to reach the correct registration, and it is commonly an iterative strategy. Below are reported some of the implemented optimization strategies.[7]
- Gradient descent
- Robbins-Monro, similar to the gradient descent, but employing an approximation of the cost function derivatives
A wider range of optimizers is also available, such as Quasi-Newton or evolutionary strategies.
Other features
The elastix software also offers other features that can be employed to speed up the registration procedure and to provide more advanced algorithms to the end-users. Some examples are the introduction of blur and Gaussian pyramid to reduce data complexity, and multi-image and multi-metric framework to deal with more complex applications.[7]
Applications
Elastix has applications mainly in the medical field, where image registration is fundamental to get comprehensive information regarding the analysed anatomical region.[12] It is widely employed in image-guided surgery, tumour monitoring, and treatment assessment.[5]
For example, in radiotherapy planning, image registration allows to correctly deliver the treatment and evaluate the obtained results. Thanks to the wide range of implemented algorithms, the use of the elastix software allows physicians and researchers to test different registration pipelines from the simplest to more complex ones, and to save the best one as a configuration file. This file and the fact that the software is completely open-source makes it easy to reproduce the work, that can help supporting the open science paradigm, and allows fast reuse on different patients data.[13]
In image-guided surgery, registration time and accuracy are critical points, considering that, during the registration, the patient is on the operating table, and the images to be registered have lower resolution compared to the target ones. In this field, the possibility to exploit elastix with high-level languages, such as OpenCL, opens to research in the usage of GPUs and other hardware accelerators.[14]
References
- ↑ "ITK | Insight Toolkit". https://itk.org/.
- ↑ Klein, S.; Staring, M.; Murphy, K.; Viergever, M.A.; Pluim, J. (January 2010). "elastix: A Toolbox for Intensity-Based Medical Image Registration". IEEE Transactions on Medical Imaging 29 (1): 196–205. doi:10.1109/TMI.2009.2035616. PMID 19923044.
- ↑ "SimpleElastix". http://simpleelastix.github.io/.
- ↑ Marstal, Kasper; Berendsen, Floris; Staring, Marius; Klein, Stefan (2016). "SimpleElastix: A User-Friendly, Multi-Lingual Library for Medical Image Registration". Proceedings of the IEEE Conference on Computer Vision and Pattern Recognition (CVPR) Workshops. pp. 134–142. https://www.cv-foundation.org/openaccess/content_cvpr_2016_workshops/w15/html/Marstal_SimpleElastix_A_User-Friendly_CVPR_2016_paper.html.
- ↑ 5.0 5.1 Zitová, Barbara; Flusser, Jan (October 2003). "Image registration methods: a survey". Image and Vision Computing 21 (11): 977–1000. doi:10.1016/S0262-8856(03)00137-9.
- ↑ 6.0 6.1 Brown, Lisa Gottesfeld (1 December 1992). "A survey of image registration techniques". ACM Computing Surveys 24 (4): 325–376. doi:10.1145/146370.146374.
- ↑ 7.0 7.1 7.2 7.3 7.4 7.5 7.6 Klein, Stefan; Staring, Marius. "elastix, the manual". https://elastix.lumc.nl/download/elastix-5.0.0-manual.pdf.
- ↑ Rueckert, D.; Sonoda, L.I.; Hayes, C.; Hill, D.L.G.; Leach, M.O.; Hawkes, D.J. (1999). "Nonrigid registration using free-form deformations: application to breast MR images". IEEE Transactions on Medical Imaging 18 (8): 712–721. doi:10.1109/42.796284. PMID 10534053.
- ↑ Davis, M.H.; Khotanzad, A.; Flamig, D.P.; Harms, S.E. (June 1997). "A physics-based coordinate transformation for 3-D image matching". IEEE Transactions on Medical Imaging 16 (3): 317–328. doi:10.1109/42.585766. PMID 9184894.
- ↑ Unser, M.; Thevenaz, P. (2000). "Optimization of mutual information for multiresolution image registration". IEEE Transactions on Image Processing 9 (12): 2083–2099. doi:10.1109/83.887976. PMID 18262946. Bibcode: 2000ITIP....9.2083U. https://infoscience.epfl.ch/record/63070/files/thevenaz0003.pdf.
- ↑ Unser, M. (1999). "Splines: a perfect fit for signal and image processing". IEEE Signal Processing Magazine 16 (6): 22–38. doi:10.1109/79.799930. Bibcode: 1999ISPM...16...22U. http://infoscience.epfl.ch/record/63064.
- ↑ Maintz, J.B.Antoine; Viergever, Max A. (March 1998). "A survey of medical image registration". Medical Image Analysis 2 (1): 1–36. doi:10.1016/S1361-8415(01)80026-8. PMID 10638851.
- ↑ Zukauskaite, Ruta; Brink, Carsten; Hansen, Christian Rønn; Bertelsen, Anders; Johansen, Jørgen; Grau, Cai; Eriksen, Jesper Grau (20 June 2016). "Open source deformable image registration system for treatment planning and recurrence CT scans". Strahlentherapie und Onkologie 192 (8): 545–551. doi:10.1007/s00066-016-0998-4. PMID 27323754.
- ↑ Shamonin, Denis (2013). "Fast parallel image registration on CPU and GPU for diagnostic classification of Alzheimer's disease". Frontiers in Neuroinformatics 7: 50. doi:10.3389/fninf.2013.00050. PMID 24474917.
External links
 |
