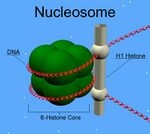
Biology:Histone H1
| linker histone H1 and H5 family | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
 | |||||||||||
| Identifiers | |||||||||||
| Symbol | Linker_histone | ||||||||||
| Pfam | PF00538 | ||||||||||
| InterPro | IPR005818 | ||||||||||
| SMART | SM00526 | ||||||||||
| SCOP2 | 1hst / SCOPe / SUPFAM | ||||||||||
| |||||||||||
Histone H1 is one of the five main histone protein families which are components of chromatin in eukaryotic cells. Though highly conserved, it is nevertheless the most variable histone in sequence across species.
Structure
Metazoan H1 proteins feature a central globular "winged helix" domain and long C- and short N-terminal tails. H1 is involved with the packing of the "beads on a string" sub-structures into a high order structure, whose details have not yet been solved.[1] H1 found in protists and bacteria, otherwise known as nucleoproteins HC1 and HC2 (Pfam PF07432, PF07382), lack the central domain and the N-terminal tail.[2]
H1 is less conserved than core histones. The globular domain is the most conserved part of H1.[3]
Function
Unlike the other histones, H1 does not make up the nucleosome "bead". Instead, it sits on top of the structure, keeping in place the DNA that has wrapped around the nucleosome. H1 is present in half the amount of the other four histones, which contribute two molecules to each nucleosome bead. In addition to binding to the nucleosome, the H1 protein binds to the "linker DNA" (approximately 20-80 nucleotides in length) region between nucleosomes, helping stabilize the zig-zagged 30 nm chromatin fiber.[4] Much has been learned about histone H1 from studies on purified chromatin fibers. Ionic extraction of linker histones from native or reconstituted chromatin promotes its unfolding under hypotonic conditions from fibers of 30 nm width to beads-on-a-string nucleosome arrays.[5][6][7]
It is uncertain whether H1 promotes a solenoid-like chromatin fiber, in which exposed linker DNA is shortened, or whether it merely promotes a change in the angle of adjacent nucleosomes, without affecting linker length[8] However, linker histones have been demonstrated to drive the compaction of chromatin fibres that had been reconstituted in vitro using synthetic DNA arrays of the strong '601' nucleosome positioning element.[9] Nuclease digestion and DNA footprinting experiments suggest that the globular domain of histone H1 localizes near the nucleosome dyad, where it protects approximately 15-30 base pairs of additional DNA.[10][11][12][13] In addition, experiments on reconstituted chromatin reveal a characteristic stem motif at the dyad in the presence of H1.[14] Despite gaps in our understanding, a general model has emerged wherein H1's globular domain closes the nucleosome by crosslinking incoming and outgoing DNA, while the tail binds to linker DNA and neutralizes its negative charge.[8][12]
Many experiments addressing H1 function have been performed on purified, processed chromatin under low-salt conditions, but H1's role in vivo is less certain. Cellular studies have shown that overexpression of H1 can cause aberrant nuclear morphology and chromatin structure, and that H1 can serve as both a positive and negative regulator of transcription, depending on the gene.[15][16][17] In Xenopus egg extracts, linker histone depletion causes ~2-fold lengthwise extension of mitotic chromosomes, while overexpression causes chromosomes to hypercompact into an inseparable mass.[18][19] Complete knockout of H1 in vivo has not been achieved in multicellular organisms due to the existence of multiple isoforms that may be present in several gene clusters, but various linker histone isoforms have been depleted to varying degrees in Tetrahymena, C. elegans, Arabidopsis, fruit fly, and mouse, resulting in various organism-specific defects in nuclear morphology, chromatin structure, DNA methylation, and/or specific gene expression.[20][21][22]
Dynamics
While most histone H1 in the nucleus is bound to chromatin, H1 molecules shuttle between chromatin regions at a fairly high rate.[23][24]
It is difficult to understand how such a dynamic protein could be a structural component of chromatin, but it has been suggested that the steady-state equilibrium within the nucleus still strongly favors association between H1 and chromatin, meaning that despite its dynamics, the vast majority of H1 at any given timepoint is chromatin bound.[25] H1 compacts and stabilizes DNA under force and during chromatin assembly, which suggests that dynamic binding of H1 may provide protection for DNA in situations where nucleosomes need to be removed.[26]
Cytoplasmic factors appear to be necessary for the dynamic exchange of histone H1 on chromatin, but these have yet to be specifically identified.[27] H1 dynamics may be mediated to some degree by O-glycosylation and phosphorylation. O-glycosylation of H1 may promote chromatin condensation and compaction. Phosphorylation during interphase has been shown to decrease H1 affinity for chromatin and may promote chromatin decondensation and active transcription. However, during mitosis phosphorylation has been shown to increase the affinity of H1 for chromosomes and therefore promote mitotic chromosome condensation.[19]
Isoforms
The H1 family in animals includes multiple H1 isoforms that can be expressed in different or overlapping tissues and developmental stages within a single organism. The reason for these multiple isoforms remains unclear, but both their evolutionary conservation from sea urchin to humans as well as significant differences in their amino acid sequences suggest that they are not functionally equivalent.[28][29][3] One isoform is histone H5, which is only found in avian erythrocytes, which are unlike mammalian erythrocytes in that they have nuclei. Another isoform is the oocyte/zygotic H1M isoform (also known as B4 or H1foo), found in sea urchins, frogs, mice, and humans, which is replaced in the embryo by somatic isoforms H1A-E, and H10 which resembles H5.[3][30][31][32] Despite having more negative charges than somatic isoforms, H1M binds with higher affinity to mitotic chromosomes in Xenopus egg extracts.[19]
Post-translational modifications
Like other histones, the histone H1 family is extensively post-translationally modified (PTMs). This includes serine and threonine phosphorylation, lysine acetylation, lysine methylation and ubiquitination.[33] These PTMs serve a variety of functions but are less well studied than the PTMs of other histones.
See also
- Nucleosome
- Histone
- Chromatin
- Linker histone H1 variants
- Other histone proteins involved in chromatin:
References
- ↑ "Crystal structure of globular domain of histone H5 and its implications for nucleosome binding". Nature 362 (6417): 219–23. March 1993. doi:10.1038/362219a0. PMID 8384699. Bibcode: 1993Natur.362..219R.
- ↑ "Origin of H1 linker histones". FASEB Journal 15 (1): 34–42. January 2001. doi:10.1096/fj.00-0237rev. PMID 11149891. https://faseb.onlinelibrary.wiley.com/doi/epdf/10.1096/fj.00-0237rev.
- ↑ 3.0 3.1 3.2 "The histone H1 family: specific members, specific functions?". Biological Chemistry 389 (4): 333–43. April 2008. doi:10.1515/BC.2008.037. PMID 18208346. https://www.degruyter.com/document/doi/10.1515/BC.2008.037/html.
- ↑ Jeon, Kwang W.; Berezney, Ronald (1995). Structural and functional organization of the nuclear matrix. Boston: Academic Press. pp. 214–7. ISBN 978-0-12-364565-4. https://archive.org/details/structuralfuncti00jeon.
- ↑ "Solenoidal model for superstructure in chromatin". Proceedings of the National Academy of Sciences of the United States of America 73 (6): 1897–901. June 1976. doi:10.1073/pnas.73.6.1897. PMID 1064861. Bibcode: 1976PNAS...73.1897F.
- ↑ "Influence of histone H1 on chromatin structure". Cell 12 (1): 101–7. September 1977. doi:10.1016/0092-8674(77)90188-X. PMID 561660. https://www.cell.com/cell/pdf/0092-8674(77)90188-X.pdf?_returnURL=https%3A%2F%2Flinkinghub.elsevier.com%2Fretrieve%2Fpii%2F009286747790188X%3Fshowall%3Dtrue.
- ↑ "Involvement of histone H1 in the organization of the nucleosome and of the salt-dependent superstructures of chromatin". The Journal of Cell Biology 83 (2 Pt 1): 403–27. November 1979. doi:10.1083/jcb.83.2.403. PMID 387806.
- ↑ 8.0 8.1 "What determines the folding of the chromatin fiber?". Proceedings of the National Academy of Sciences of the United States of America 93 (20): 10548–55. October 1996. doi:10.1073/pnas.93.20.10548. PMID 8855215. Bibcode: 1996PNAS...9310548V.
- ↑ "Nucleosome repeat length and linker histone stoichiometry determine chromatin fiber structure". Proceedings of the National Academy of Sciences of the United States of America 105 (26): 8872–7. July 2008. doi:10.1073/pnas.0802336105. PMID 18583476. Bibcode: 2008PNAS..105.8872R.
- ↑ "Heterogeneity of chromatin subunits in vitro and location of histone H1". Nucleic Acids Research 3 (2): 477–92. February 1976. doi:10.1093/nar/3.2.477. PMID 1257057.
- ↑ "Removal of histone H1 exposes a fifty base pair DNA segment between nucleosomes". Biochemistry 15 (15): 3307–14. July 1976. doi:10.1021/bi00660a022. PMID 952859. https://pubs.acs.org/doi/abs/10.1021/bi00660a022.
- ↑ 12.0 12.1 "The structure of histone H1 and its location in chromatin". Nature 288 (5792): 675–9. December 1980. doi:10.1038/288675a0. PMID 7453800. Bibcode: 1980Natur.288..675A. https://pubmed.ncbi.nlm.nih.gov/7453800/.
- ↑ "Footprinting of linker histones H5 and H1 on the nucleosome". The EMBO Journal 7 (12): 3685–91. December 1988. doi:10.1002/j.1460-2075.1988.tb03250.x. PMID 3208745.
- ↑ "Nucleosomes, linker DNA, and linker histone form a unique structural motif that directs the higher-order folding and compaction of chromatin". Proceedings of the National Academy of Sciences of the United States of America 95 (24): 14173–8. November 1998. doi:10.1073/pnas.95.24.14173. PMID 9826673. Bibcode: 1998PNAS...9514173B.
- ↑ "The maternal histone H1 variant, H1M (B4 protein), is the predominant H1 histone in Xenopus pregastrula embryos". Developmental Biology 161 (2): 425–39. February 1994. doi:10.1006/dbio.1994.1042. PMID 8313993.
- ↑ "Differential effect of H1 variant overexpression on cell cycle progression and gene expression". Nucleic Acids Research 24 (3): 486–93. February 1996. doi:10.1093/nar/24.3.486. PMID 8602362.
- ↑ "Effects of H1 histone variant overexpression on chromatin structure". The Journal of Biological Chemistry 274 (53): 37950–6. December 1999. doi:10.1074/jbc.274.53.37950. PMID 10608862.
- ↑ "Histone H1 is essential for mitotic chromosome architecture and segregation in Xenopus laevis egg extracts". The Journal of Cell Biology 169 (6): 859–69. June 2005. doi:10.1083/jcb.200503031. PMID 15967810.
- ↑ 19.0 19.1 19.2 "Functional comparison of H1 histones in Xenopus reveals isoform-specific regulation by Cdk1 and RanGTP". Current Biology 20 (11): 1048–52. June 2010. doi:10.1016/j.cub.2010.04.025. PMID 20471264.
- ↑ "Linker histones are not essential and affect chromatin condensation in vivo". Cell 82 (1): 47–56. July 1995. doi:10.1016/0092-8674(95)90051-9. PMID 7606784.
- ↑ "A single histone H1 isoform (H1.1) is essential for chromatin silencing and germline development in Caenorhabditis elegans". Development 128 (7): 1069–80. April 2001. doi:10.1242/dev.128.7.1069. PMID 11245572. https://journals.biologists.com/dev/article/128/7/1069/41594/A-single-histone-H1-isoform-H1-1-is-essential-for.
- ↑ "Linker histone H1 is essential for Drosophila development, the establishment of pericentric heterochromatin, and a normal polytene chromosome structure". Genes & Development 23 (4): 452–65. February 2009. doi:10.1101/gad.1749309. PMID 19196654.
- ↑ "Dynamic binding of histone H1 to chromatin in living cells". Nature 408 (6814): 877–81. December 2000. doi:10.1038/35048610. PMID 11130729. Bibcode: 2000Natur.408..877M.
- ↑ "Condensed mitotic chromatin is accessible to transcription factors and chromatin structural proteins". The Journal of Cell Biology 168 (1): 41–54. January 2005. doi:10.1083/jcb.200407182. PMID 15623580.
- ↑ "The dynamics of histone H1 function in chromatin". Molecular Cell 17 (5): 617–20. March 2005. doi:10.1016/j.molcel.2005.02.019. PMID 15749012.
- ↑ "Histone H1 compacts DNA under force and during chromatin assembly". Molecular Biology of the Cell 23 (24): 4864–71. December 2012. doi:10.1091/mbc.E12-07-0518. PMID 23097493.
- ↑ Cimini, Daniela, ed (September 2010). "Xenopus egg extracts increase dynamics of histone H1 on sperm chromatin". PLOS ONE 5 (9): e13111. doi:10.1371/journal.pone.0013111. PMID 20927327. Bibcode: 2010PLoSO...513111F.
- ↑ "Somatic linker histones cause loss of mesodermal competence in Xenopus". Nature 389 (6649): 395–9. September 1997. doi:10.1038/38755. PMID 9311783. Bibcode: 1997Natur.389..395S.
- ↑ "Histone H1 variants differentially inhibit DNA replication through an affinity for chromatin mediated by their carboxyl-terminal domains". Gene 292 (1–2): 173–81. June 2002. doi:10.1016/S0378-1119(02)00675-3. PMID 12119111.
- ↑ "Histone H1 diversity: bridging regulatory signals to linker histone function". Gene 271 (1): 1–12. June 2001. doi:10.1016/S0378-1119(01)00495-4. PMID 11410360. https://www.sciencedirect.com/science/article/abs/pii/S0378111901004954.
- ↑ "Cracking the enigmatic linker histone code". Journal of Biochemistry 143 (3): 287–93. March 2008. doi:10.1093/jb/mvn013. PMID 18234717. https://academic.oup.com/jb/article-abstract/143/3/287/2182517?redirectedFrom=fulltext&login=false.
- ↑ "Histone H1 and its isoforms: contribution to chromatin structure and function". Gene 431 (1–2): 1–12. February 2009. doi:10.1016/j.gene.2008.11.003. PMID 19059319. https://www.sciencedirect.com/science/article/abs/pii/S0378111908005684.
- ↑ "H1 histones: current perspectives and challenges". Nucleic Acids Research 41 (21): 9593–609. November 2013. doi:10.1093/nar/gkt700. PMID 23945933.
 |