Biology:Chromosome 22
| Chromosome 22 | |
|---|---|
 Human chromosome 22 pair after G-banding. One is from mother, one is from father. | |
 Chromosome 22 pair in human male karyogram. | |
| Features | |
| Length (bp) | 51,324,926 bp (CHM13) |
| No. of genes | 417 (CCDS)[1] |
| Type | Autosome |
| Centromere position | Acrocentric[2] (15.0 Mbp[3]) |
| Complete gene lists | |
| CCDS | Gene list |
| HGNC | Gene list |
| UniProt | Gene list |
| NCBI | Gene list |
| External map viewers | |
| Ensembl | Chromosome 22 |
| Entrez | Chromosome 22 |
| NCBI | Chromosome 22 |
| UCSC | Chromosome 22 |
| Full DNA sequences | |
| RefSeq | NC_000022 (FASTA) |
| GenBank | CM000684 (FASTA) |
Chromosome 22 is one of the 23 pairs of chromosomes in human cells. Humans normally have two copies of chromosome 22 in each cell. Chromosome 22 is the second smallest human chromosome, spanning about 51 million DNA base pairs and representing between 1.5 and 2% of the total DNA in cells.
In 1999, researchers working on the Human Genome Project announced they had determined the sequence of base pairs that make up this chromosome. Chromosome 22 was the first human chromosome to be fully sequenced.[4]
Human chromosomes are numbered by their apparent size in the karyotype, with Chromosome 1 being the largest and Chromosome 22 having originally been identified as the smallest. However, genome sequencing has revealed that Chromosome 21 is actually smaller than Chromosome 22.
Genes
Number of genes
The following are some of the gene count estimates of human chromosome 22. Because researchers use different approaches to genome annotation, their predictions of the number of genes on each chromosome varies (for technical details, see gene prediction). Among various projects, the collaborative consensus coding sequence project (CCDS) takes an extremely conservative strategy. So CCDS's gene number prediction represents a lower bound on the total number of human protein-coding genes.[5]
| Estimated by | Protein-coding genes | Non-coding RNA genes | Pseudogenes | Source | Release date |
|---|---|---|---|---|---|
| CCDS | 417 | — | — | [1] | 2016-09-08 |
| HGNC | 424 | 161 | 295 | [6] | 2019-07-08 |
| Ensembl | 489 | 515 | 325 | [7] | 2017-03-29 |
| UniProt | 496 | — | — | [8] | 2018-02-28 |
| NCBI | 474 | 392 | 379 | [9][10][11] | 2017-05-19 |
Gene list
The following is a partial list of genes on human chromosome 22. For complete list, see the link in the infobox on the right.
| Locus | Gene | Description | Condition |
| Template:Locus | IGL@ | Asymmetric crying facies (Cayler cardiofacial syndrome) | |
| Template:Locus | TBX1 | T-box 1 | |
| Template:Locus | RTN4R | Reticulon 4 receptor | Schizophrenia |
| Template:Locus | COMT | catechol-O-methyltransferase gene | |
| Template:Locus | NEFH | neurofilament, heavy polypeptide 200kDa | |
| Template:Locus[12] | CHEK2 | CHK2 checkpoint homolog (S. pombe) | |
| Template:Locus | NF2 | neurofibromin 2 | bilateral acoustic neuroma |
| Template:Locus | SOX10 | SRY (sex determining region Y)-box 10 | |
| Template:Locus | APOL1 | Apolipoprotein L1 | |
| Template:Locus | EP300 | E1A binding protein p300 | |
| Template:Locus | WNT7B | Wingless-type MMTV integration site family, member 7B | 22q13 deletion syndrome |
| Template:Locus | SHANK3 | SH3 and multiple ankyrin repeat domains 3 | 22q13 deletion syndrome |
| Template:Locus | SULT4A1 | sulfotransferase family 4A, member 1 | 22q13 deletion syndrome |
| Template:Locus | PARVB | parvin beta (cytoskeleton organization and cell adhesion) | 22q13 deletion syndrome |
Diseases and disorders
The following diseases are some of those related to genes on chromosome 22:
- Amyotrophic lateral sclerosis
- Breast cancer
- Cat eye syndrome
- Chronic myeloid leukemia
- DiGeorge Syndrome
- Desmoplastic small round cell tumor
- 22q11.2 distal deletion syndrome
- 22q13 deletion syndrome or Phelan-McDermid syndrome
- Emanuel syndrome
- Ewing sarcoma
- Focal Segmental Glomerulosclerosis
- Li-Fraumeni syndrome
- Metachromatic leukodystrophy
- Methemoglobinemia
- Neurofibromatosis type 2
- Opitz G/BBB syndrome
- Renal Medullary Carcinoma
- Rubinstein-Taybi syndrome
- Waardenburg syndrome
- Schizophrenia[13]
Chromosomal conditions
The following conditions are caused by changes in the structure or number of copies of chromosome 22:
- 22q11.2 deletion syndrome: Most people with 22q11.2 deletion syndrome are missing about 3 million base pairs on one copy of chromosome 22 in each cell. The deletion occurs near the middle of the chromosome at a location designated as q11.2. This region contains about 30 genes, but many of these genes have not been well characterized. A small percentage of affected individuals have shorter deletions in the same region.
The loss of one particular gene, TBX1, is thought to be responsible for many of the characteristic features of 22q11.2 deletion syndrome, such as heart defects, an opening in the roof of the mouth (a cleft palate), distinctive facial features, and low calcium levels. A loss of this gene does not appear to cause learning disabilities, however. Other genes in the deleted region are also likely to contribute to the signs and symptoms of 22q11.2 deletion syndrome. - 22q11.2 distal deletion syndrome
- 22q13 deletion syndrome
- Other chromosomal conditions: Other changes in the number or structure of chromosome 22 can have a variety of effects, including intellectual disability, delayed development, physical abnormalities, and other medical problems. These changes include an extra piece of chromosome 22 in each cell (partial trisomy), a missing segment of the chromosome in each cell (partial monosomy), and a circular structure called ring chromosome 22 that is caused by the breakage and reattachment of both ends of the chromosome.
- Cat-eye syndrome is a rare disorder most often caused by a chromosomal change called an inverted duplicated 22. A small extra chromosome is made up of genetic material from chromosome 22 that has been abnormally duplicated (copied). The extra genetic material causes the characteristic signs and symptoms of cat-eye syndrome, including an eye abnormality called ocular iris coloboma (a gap or split in the colored part of the eye), small skin tags or pits in front of the ear, heart defects, kidney problems, and, in some cases, delayed development.
- A rearrangement (translocation) of genetic material between chromosomes 9 and 22 is associated with several types of blood cancer (leukemia). This chromosomal abnormality, which is commonly called the Philadelphia chromosome, is found only in cancer cells. The Philadelphia chromosome has been identified in most cases of a slowly progressing form of blood cancer called chronic myeloid leukemia, or CML. It also has been found in some cases of more rapidly progressing blood cancers (acute leukemias). The presence of the Philadelphia chromosome can help predict how the cancer will progress and provides a target for molecular therapies.
- Emanuel syndrome is a translocation of chromosomes 11 and 22. Originally known as Supernumerary der (22) Syndrome, it occurs when an individual has an extra chromosome composed of pieces of the 11th and 22nd chromosomes.
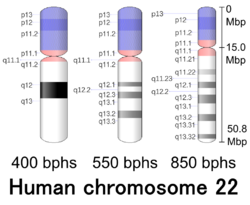
Cytogenetic band
| Chr. | Arm[18] | Band[19] | ISCN start[20] |
ISCN stop[20] |
Basepair start |
Basepair stop |
Stain[21] | Density |
|---|---|---|---|---|---|---|---|---|
| 22 | p | 13 | 0 | 260 | 1 | 4,300,000 | gvar | |
| 22 | p | 12 | 260 | 576 | 4,300,001 | 9,400,000 | stalk | |
| 22 | p | 11.2 | 576 | 836 | 9,400,001 | 13,700,000 | gvar | |
| 22 | p | 11.1 | 836 | 1015 | 13,700,001 | 15,000,000 | acen | |
| 22 | q | 11.1 | 1015 | 1234 | 15,000,001 | 17,400,000 | acen | |
| 22 | q | 11.21 | 1234 | 1563 | 17,400,001 | 21,700,000 | gneg | |
| 22 | q | 11.22 | 1563 | 1700 | 21,700,001 | 23,100,000 | gpos | 25 |
| 22 | q | 11.23 | 1700 | 1878 | 23,100,001 | 25,500,000 | gneg | |
| 22 | q | 12.1 | 1878 | 2029 | 25,500,001 | 29,200,000 | gpos | 50 |
| 22 | q | 12.2 | 2029 | 2194 | 29,200,001 | 31,800,000 | gneg | |
| 22 | q | 12.3 | 2194 | 2413 | 31,800,001 | 37,200,000 | gpos | 50 |
| 22 | q | 13.1 | 2413 | 2687 | 37,200,001 | 40,600,000 | gneg | |
| 22 | q | 13.2 | 2687 | 2852 | 40,600,001 | 43,800,000 | gpos | 50 |
| 22 | q | 13.31 | 2852 | 3181 | 43,800,001 | 48,100,000 | gneg | |
| 22 | q | 13.32 | 3181 | 3290 | 48,100,001 | 49,100,000 | gpos | 50 |
| 22 | q | 13.33 | 3290 | 3400 | 49,100,001 | 50,818,468 | gneg |
References
- ↑ 1.0 1.1 "Search results - 22[CHR] AND "Homo sapiens"[Organism] AND ("has ccds"[Properties] AND alive[prop]) - Gene". 2016-09-08. https://www.ncbi.nlm.nih.gov/gene?term=22%5BChr%5D%20AND%20%22Homo%20sapiens%22%5BOrganism%5D%20AND%20%28%22has%20ccds%22%5BProperties%5D%20AND%20alive%5Bprop%5D%29&cmd=DetailsSearch.
- ↑ Tom Strachan; Andrew Read (2 April 2010). Human Molecular Genetics. Garland Science. p. 45. ISBN 978-1-136-84407-2. https://books.google.com/books?id=dSwWBAAAQBAJ&pg=PA45.
- ↑ 3.0 3.1 3.2 Genome Decoration Page, NCBI. Ideogram data for Homo sapience (850 bphs, Assembly GRCh38.p3). Last update 2014-06-03. Retrieved 2017-04-26.
- ↑ Mayor, Susan (1999). "First human chromosome is sequenced". BMJ (BMJ Group) 319 (7223): 1453. doi:10.1136/bmj.319.7223.1453a. PMID 10582915.
- ↑ Pertea M, Salzberg SL (2010). "Between a chicken and a grape: estimating the number of human genes.". Genome Biol 11 (5): 206. doi:10.1186/gb-2010-11-5-206. PMID 20441615.
- ↑ "Statistics & Downloads for chromosome 22". 2019-07-08. https://www.genenames.org/cgi-bin/statistics?c=22.
- ↑ "Chromosome 22: Chromosome summary - Homo sapiens". 2017-03-29. http://mar2017.archive.ensembl.org/Homo_sapiens/Location/Chromosome?r=22.
- ↑ "Human chromosome 22: entries, gene names and cross-references to MIM". 2018-02-28. https://www.uniprot.org/docs/humchr22.txt.
- ↑ "Search results - 22[CHR] AND "Homo sapiens"[Organism] AND ("genetype protein coding"[Properties] AND alive[prop]) - Gene". 2017-05-19. https://www.ncbi.nlm.nih.gov/gene?term=22%5BCHR%5D%20AND%20%22Homo%20sapiens%22%5BOrganism%5D%20AND%20%28%22genetype%20protein%20coding%22%5BProperties%5D%20AND%20alive%5Bprop%5D%29&cmd=DetailsSearch.
- ↑ "Search results - 22[CHR] AND "Homo sapiens"[Organism] AND ( ("genetype miscrna"[Properties] OR "genetype ncrna"[Properties] OR "genetype rrna"[Properties] OR "genetype trna"[Properties] OR "genetype scrna"[Properties] OR "genetype snrna"[Properties] OR "genetype snorna"[Properties]) NOT "genetype protein coding"[Properties] AND alive[prop]) - Gene". 2017-05-19. https://www.ncbi.nlm.nih.gov/gene?term=22%5BCHR%5D%20AND%20%22Homo%20sapiens%22%5BOrganism%5D%20AND%20%28%28%22genetype%20miscrna%22%5BProperties%5D%20OR%20%22genetype%20ncrna%22%5BProperties%5D%20OR%20%22genetype%20rrna%22%5BProperties%5D%20OR%20%22genetype%20trna%22%5BProperties%5D%20OR%20%22genetype%20scrna%22%5BProperties%5D%20OR%20%22genetype%20snrna%22%5BProperties%5D%20OR%20%22genetype%20snorna%22%5BProperties%5D%29%20NOT%20%22genetype%20protein%20coding%22%5BProperties%5D%20AND%20alive%5Bprop%5D%29&cmd=DetailsSearch.
- ↑ "Search results - 22[CHR] AND "Homo sapiens"[Organism] AND ("genetype pseudo"[Properties] AND alive[prop]) - Gene". 2017-05-19. https://www.ncbi.nlm.nih.gov/gene?term=22%5BCHR%5D%20AND%20%22Homo%20sapiens%22%5BOrganism%5D%20AND%20%28%22genetype%20pseudo%22%5BProperties%5D%20AND%20alive%5Bprop%5D%29&cmd=DetailsSearch.
- ↑ Beck, Megan; Peterson, Jess F.; McConnell, Juliann; McGuire, Marianne; Asato, Miya; Losee, Joseph E.; Surti, Urvashi; Madan-Khetarpal, Suneeta et al. (May 2015). "Craniofacial abnormalities and developmental delay in two families with overlapping 22q12.1 microdeletions involving the gene". American Journal of Medical Genetics Part A 167 (5): 1047–1053. doi:10.1002/ajmg.a.36839. PMID 25810350. https://escholarship.org/uc/item/0vx445vv#page-1.
- ↑ "Genetic variation in the 22q11 locus and susceptibility to schizophrenia". Proc. Natl. Acad. Sci. U.S.A. 99 (26): 16859–64. December 2002. doi:10.1073/pnas.232186099. PMID 12477929. Bibcode: 2002PNAS...9916859L.
- ↑ Genome Decoration Page, NCBI. Ideogram data for Homo sapience (400 bphs, Assembly GRCh38.p3). Last update 2014-03-04. Retrieved 2017-04-26.
- ↑ Genome Decoration Page, NCBI. Ideogram data for Homo sapience (550 bphs, Assembly GRCh38.p3). Last update 2015-08-11. Retrieved 2017-04-26.
- ↑ International Standing Committee on Human Cytogenetic Nomenclature (2013). ISCN 2013: An International System for Human Cytogenetic Nomenclature (2013). Karger Medical and Scientific Publishers. ISBN 978-3-318-02253-7. https://books.google.com/books?id=lGCLrh0DIwEC.
- ↑ Sethakulvichai, W.; Manitpornsut, S.; Wiboonrat, M.; Lilakiatsakun, W.; Assawamakin, A.; Tongsima, S. (2012). "Estimation of band level resolutions of human chromosome images". 2012 Ninth International Conference on Computer Science and Software Engineering (JCSSE). pp. 276–282. doi:10.1109/JCSSE.2012.6261965. ISBN 978-1-4673-1921-8. https://www.researchgate.net/publication/261304470.
- ↑ "p": Short arm; "q": Long arm.
- ↑ For cytogenetic banding nomenclature, see article locus.
- ↑ 20.0 20.1 These values (ISCN start/stop) are based on the length of bands/ideograms from the ISCN book, An International System for Human Cytogenetic Nomenclature (2013). Arbitrary unit.
- ↑ gpos: Region which is positively stained by G banding, generally AT-rich and gene poor; gneg: Region which is negatively stained by G banding, generally CG-rich and gene rich; acen Centromere. var: Variable region; stalk: Stalk.
Further reading
- "The DNA sequence of human chromosome 22". Nature 402 (6761): 489–95. 1999. doi:10.1038/990031. PMID 10591208. Bibcode: 1999Natur.402..489D.
- Gilbert F (1998). "Disease genes and chromosomes: disease maps of the human genome. Chromosome 22". Genet Test 2 (1): 89–97. doi:10.1089/gte.1998.2.89. PMID 10464604.
- "Philadelphia chromosome-positive leukemias: from basic mechanisms to molecular therapeutics". Ann Intern Med 138 (10): 819–30. 2003. doi:10.7326/0003-4819-138-10-200305200-00010. PMID 12755554.
- "22q11 DS: genomic mechanisms and gene function in DiGeorge/velocardiofacial syndrome". Int J Dev Neurosci 20 (3–5): 407–19. 2002. doi:10.1016/S0736-5748(02)00050-3. PMID 12175881.
- "Genomic disorders on 22q11". Am J Hum Genet 70 (5): 1077–88. 2002. doi:10.1086/340363. PMID 11925570.
- "The Philadelphia story: the 22q11.2 deletion: report on 250 patients". Genet Couns 10 (1): 11–24. 1999. PMID 10191425.
- "The transcriptional activity of human Chromosome 22". Genes Dev 17 (4): 529–40. 2003. doi:10.1101/gad.1055203. PMID 12600945.
- "Molecular characterisation of the 22q13 deletion syndrome supports the role of haploinsufficiency of SHANK3/PROSASP2 in the major neurological symptoms". J Med Genet 40 (8): 575–584. 2003. doi:10.1136/jmg.40.8.575. PMID 12920066.
External links
- National Institutes of Health. "Chromosome 22". Genetics Home Reference. http://ghr.nlm.nih.gov/chromosome=22.
- "Chromosome 22". http://web.ornl.gov/sci/techresources/Human_Genome/posters/chromosome/chromo22.shtml.
 |