Biology:RAD51
 Generic protein structure example |
DNA repair protein RAD51 homolog 1 is a protein encoded by the gene RAD51. The enzyme encoded by this gene is a member of the RAD51 protein family which assists in repair of DNA double strand breaks. RAD51 family members are homologous to the bacterial RecA, Archaeal RadA and yeast Rad51.[1][2] The protein is highly conserved in most eukaryotes, from yeast to humans.[3]
The name RAD51 derives from RADiation sensitive protein 51.[4]
Variants
Two alternatively spliced transcript variants of this gene, which encode distinct proteins, have been reported. Transcript variants utilizing alternative polyA signals exist.
Family
In mammals, seven recA-like genes have been identified: Rad51, Rad51L1/B, Rad51L2/C, Rad51L3/D, XRCC2, XRCC3, and DMC1/Lim15.[5] All of these proteins, with the exception of meiosis-specific DMC1, are essential for development in mammals. Rad51 is a member of the RecA-like NTPases.
Function
In humans, RAD51 is a 339-amino acid protein that plays a major role in homologous recombination of DNA during double strand break repair. In this process, an ATP dependent DNA strand exchange takes place in which a template strand invades base-paired strands of homologous DNA molecules. RAD51 is involved in the search for homology and strand pairing stages of the process.
Unlike other proteins involved in DNA metabolism, the RecA/Rad51 family forms a helical nucleoprotein filament on DNA.[6]
This protein can interact with the ssDNA-binding protein RPA, BRCA2, PALB2[7] and RAD52.
The structural basis for Rad51 filament formation and its functional mechanism still remain poorly understood. However, recent studies using fluorescent labeled Rad51[8] have indicated that Rad51 fragments elongate via multiple nucleation events followed by growth, with the total fragment terminating when it reaches about 2 μm in length. Disassociation of Rad51 from dsDNA, however, is slow and incomplete, suggesting that there is a separate mechanism that accomplishes this.
RAD51 expression in cancer
In eukaryotes, RAD51 protein has a central role in homologous recombinational repair. RAD51 catalyses strand transfer between a broken sequence and its undamaged homologue to allow re-synthesis of the damaged region (see homologous recombination models).
Numerous studies report that RAD51 is over-expressed in different cancers (see Table 1). In many of these studies, elevated expression of RAD51 is correlated with decreased patient survival. There are also some reports of under-expression of RAD51 in cancers (see Table 1).
Where RAD51 expression was measured in conjunction with BRCA1 expression, an inverse correlation was found.[9][10] This was interpreted as selection for increased RAD51 expression and thus increased homologous recombinational repair (HRR) (by the HRR RAD52-RAD51 back-up pathway[11]) to compensate for the added DNA damages remaining when BRCA1 was deficient.[9][10][12]
Many cancers have epigenetic deficiencies in various DNA repair genes (see Frequencies of epimutations in DNA repair genes in cancers), likely causing increased unrepaired DNA damages. The over expression of RAD51 seen in many cancers may reflect compensatory RAD51 over expression (as in BRCA1 deficiency) and increased HRR to at least partially deal with such excess DNA damages.
Under-expression of RAD51 would itself lead to increased unrepaired DNA damages. Replication errors past these damages (see translesion synthesis), would lead to increased mutations and cancer.
| Cancer | Over or Under expression | Frequency of altered expression | Evaluation method | Ref. |
|---|---|---|---|---|
| Breast cancer (invasive ductal) | Over-expression | - | Immunohistochemistry | [9] |
| Breast cancer (BRCA1 deficient) | Over-expression | - | messenger RNA | [10] |
| Breast cancer (progesteron receptor negative) | Over-expression | - | messenger RNA | [13] |
| Breast cancer | Under-expression | 30% | Immunohistochemistry | [14] |
| Pancreatic cancer | Over-expression | 74% | Immunohistochemistry | [15] |
| Pancreatic cancer | Over-expression | 66% | Immunohistochemistry | [16] |
| Head and neck squamous cancers | Over-expression | 75% | Immunohistochemistry | [17] |
| Prostate cancer | Over-expression | 33% | Immunohistochemistry | [18] |
| Non-small-cell lung cancer | Over-expression | 29% | Immunohistochemistry | [19] |
| Soft tissue sarcoma | Over-expression | 95% | Immunohistochemistry | [20] |
| Esophageal squamous cell cancer | Over-expression | 47% | Immunohistochemistry | [21] |
| Renal cell carcinoma | Under-expression | 100% | Western (protein) blotting and mRNA | [22] |
In double-strand break repair
Double-strand break (DSB) repair by homologous recombination is initiated by 5' to 3' strand resection (DSB resection). In humans, the DNA2 nuclease cuts back the 5'-to-3' strand at the DSB to generate a 3' single-strand DNA overhang strand.[23][24]
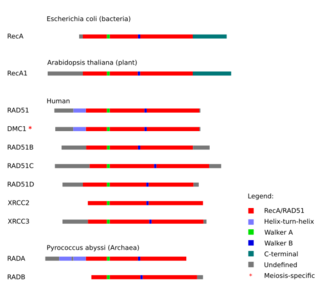
A number of paralogs (see Figure) of RAD51 are essential for RAD51 protein recruitment or stabilization at damage sites in vertebrates.
In vertebrates and plants, five paralogs of RAD51 are expressed in somatic cells, including RAD51B (RAD51L1), RAD51C (RAD51L2), RAD51D (RAD51L3), XRCC2 and XRCC3. They each share about 25% amino acid sequence identity with RAD51 and with each other.[25]
Outside of plants and vertebrates, a much broader diversity of Rad51 recombinase paralog proteins exists. In budding yeast, Saccharomyces cerevisiae, the paralogs Rad55 and Rad57 are present, which form a complex that associates with yeast Rad51 to ssDNA. The recombinase paralog rfs-1 is found in the round worm Caenorhabditis elegans, where it is not essential for homologous recombination. Among archaea the RadB and RadC recombinase paralogs are found in many organisms belonging to Euryarchaeota while a broader diversity of related recombinase paralogs seem to be found in the Crenarchaea including Ral1, Ral2, Ral3, RadC, RadC1, and RadC2.
The RAD51 paralogs contribute to efficient DNA double-strand break repair by homologous recombination and depletion of any paralog often results in significant decreases in homologous recombination frequency.[26]
The paralogs form two identified complexes: BCDX2 (RAD51B-RAD51C-RAD51D-XRCC2) and CX3 (RAD51C-XRCC3). These two complexes act at two different stages of homologous recombinational DNA repair. The BCDX2 complex is responsible for RAD51 recruitment or stabilization at damage sites.[26] The BCDX2 complex appears to act by facilitating the assembly or stability of the RAD51 nucleoprotein filament. The CX3 complex acts downstream of RAD51 recruitment to damage sites.[26]
Another complex, the BRCA1-PALB2-BRCA2 complex, and the RAD51 paralogs cooperate to load RAD51 onto ssDNA coated with RPA to form the essential recombination intermediate, the RAD51-ssDNA filament.[27]
In mice and humans, the BRCA2 complex primarily mediates orderly assembly of RAD51 on ssDNA, the form that is active for homologous pairing and strand invasion.[28] BRCA2 also redirects RAD51 from dsDNA and prevents dissociation from ssDNA.[28] However, in the presence of a BRCA2 mutation, human RAD52 can mediate RAD51 assembly on ssDNA and substitute for BRCA2 in homologous recombinational DNA repair,[29] though with lower efficiency than BRCA2.
Further steps are detailed in the article Homologous recombination.
Meiosis
Rad51 has a crucial function in meiotic prophase in mice and its loss leads to depletion of late prophase I spermatocytes.[30]
During meiosis, the two recombinases, Rad51 and Dmc1, interact with single-stranded DNA to form specialized filaments that are adapted for facilitating recombination between homologous chromosomes. Both Rad51 and Dmc1 have an intrinsic ability to self-aggregate.[31] The presence of Dmc1 stabilizes the adjacent Rad51 filaments suggesting that cross-talk between these two recombinases may affect their biochemical properties.
Chemotherapy and aging
In aged and chemotherapy treated females, oocytes and follicles are depleted by apoptosis (programmed cell death) leading to ovarian failure. DNA damage-induced oocyte apoptosis depends on the efficiency of the DNA repair machinery that in turn declines with age. Survival of oocytes following chemotherapy or aging can be enhanced by increased expression of Rad51.[32] The Rad51-induced oocyte resistance to apoptosis is likely due to Rad51’s central role in homologous recombinational repair of DNA damage.
MicroRNA control of RAD51 expression
In mammals, microRNAs (miRNAs) regulate about 60% of the transcriptional activity of protein-encoding genes.[33] Some miRNAs also undergo methylation-associated silencing in cancer cells.[34][35] If a repressive miRNA is silenced by hypermethylation or deletion, then a gene it is targeting becomes over-expressed.
At least eight miRNAs have been identified that repress RAD51 expression, and five of them appear to be important in cancer. For instance, in triple negative breast cancers (TNBC), over-expression of miR-155 occurs together with repression of RAD51.[36] Further tests directly showed that transfecting breast cancer cells with a vector over-expressing miR-155 represses RAD51, causing decreased homologous recombination and increased sensitivity to ionizing radiation.[36]
Four further miRNAs that repress RAD51 (miR-148b* and miR-193b*,[37] miR-506,[38] and miR-34a[39]) are under-expressed in cancers, presumably leading to induction of RAD51.
Under-expression of miR-148b* and miR-193b* cause an observed induction of RAD51 expression.[37] Deletions of 148b* and miR-193b* in serous ovarian tumors correlate with increased incidences[spelling?] of (possibly carcinogenic) losses of heterozygosity (LOH). This excess LOH was thought to be due to excess recombination caused by induced expression of RAD51.[37]
Under-expression of miR-506 is associated with early time to recurrence (and reduced survival) for epithelial ovarian cancer patients.[40]
Methylation of the promoter of miR-34a, resulting in under-expression of miR-34a, is observed in 79% of prostate cancers and 63% of primary melanomas.[41] Under-expressed levels of miR-34a are also seen in 63% of non-small cell lung cancers,[42] and 36% of colon cancers.[43] miR-34a is also generally under-expressed in primary neuroblastoma tumors.[44]
Table 2 summarizes these five microRNAs, their over or under expression, and the cancers in which their altered expression was noted to occur.
| MicroRNA | miRNA Over/Under expression | Cancer | Ref. |
|---|---|---|---|
| miR-155 | Over-expression | Triple negative breast cancer | [36] |
| miR-148b* | Under-expression | Ovarian cancer | [37] |
| miR-193b* | Under-expression | Ovarian cancer | [37] |
| miR-506 | Under-expression | Ovarian cancer | [40] |
| miR-34a | Under-expression | Prostate, Melanoma | [41] |
| Non-small cell lung cancer | [42] | ||
| Colon cancer | [43] | ||
| Neuroblastoma | [44] |
The information summarized in Table 2 suggests that under-expression of microRNAs (causing induction of RAD51) occurs frequently in cancers. Over-expression of a microRNA that causes repression of RAD51 appears to be less frequent. The data in Table 1 (above) indicates that, in general, over-expression of RAD51 is more frequent in cancers than under-expression.
Three other microRNAs were identified, by various criteria, as likely to repress RAD51 (miR-96,[45] miR-203,[46] and miR-103/107[47]). These microRNAs were then tested by over-expressing them in cells in vitro, and they were found to indeed repress RAD51. This repression was generally associated with decreased HR and increased sensitivity of the cells to DNA damaging agents.
Pathology
This protein is also found to interact with PALB2[7] and BRCA2, which may be important for the cellular response to DNA damage. BRCA2 is shown to regulate both the intracellular localization and DNA-binding ability of this protein. Loss of these controls following BRCA2 inactivation may be a key event leading to genomic instability and tumorigenesis.[48]
Several alterations of the Rad51 gene have been associated with an increased risk of developing breast cancer. The breast cancer susceptibility protein BRCA2 and PALB2 controls the function of Rad51 in the pathway for DNA repair by homologous recombination.[7][49] In addition to the data listed in Table 1, increased RAD51 expression levels have been identified in metastatic canine mammary carcinoma, indicating that genomic instability plays an important role in the carcinogenesis of this tumor type.[50][51][52][53]
Fanconi anemia
Fanconi anemia (FA) is a hereditary condition characterized by cellular hypersensitivity to DNA cross-linking agents. A dominant negative mutation in the Rad51 gene has been reported to give rise to an FA-like phenotype with features of mental retardation.[54][55] This report included evidence that Rad51-mediated homologous recombinational repair likely has an important role in neurodevelopment.
Interactions
RAD51 has been shown to interact with:
- Abl gene,[56]
- Ataxia telangiectasia mutated,[56]
- BARD1,[57]
- BRCA1,[57][58][59][60]
- BRCA2,[49][57][58][61][62][63][64][65][66][67][68][69][70]
- BRCC3,[57]
- BRE,[57]
- Bloom syndrome protein,[71]
- DMC1,[72]
- RAD54,[73]
- P53[57][74][75]
- RAD52,[56]
- RAD54B,[76] and
- UBE2I.[77][78]
References
- ↑ "Rad51 protein involved in repair and recombination in S. cerevisiae is a RecA-like protein". Cell 69 (3): 457–70. May 1992. doi:10.1016/0092-8674(92)90447-K. PMID 1581961.
- ↑ "RadA protein is an archaeal RecA protein homolog that catalyzes DNA strand exchange". Genes & Development 12 (9): 1248–53. May 1998. doi:10.1101/gad.12.9.1248. PMID 9573041.
- ↑ "Cloning of human, mouse and fission yeast recombination genes homologous to RAD51 and recA". Nature Genetics 4 (3): 239–43. July 1993. doi:10.1038/ng0793-239. PMID 8358431.
- ↑ Khoo, Kelvin H. P.; Jolly, Hayley R.; Able, Jason A. (2008). "The RAD51 gene family in bread wheat is highly conserved across eukaryotes, with RAD51A upregulated during early meiosis". Functional Plant Biology 35 (12): 1267–1277. doi:10.1071/fp08203. ISSN 1445-4408. PMID 32688873. http://dx.doi.org/10.1071/fp08203.
- ↑ "Role of recA/RAD51 family proteins in mammals". Acta Medica Okayama 59 (1): 1–9. February 2005. doi:10.18926/AMO/31987. PMID 15902993.
- ↑ "The Rad51/RadA N-terminal domain activates nucleoprotein filament ATPase activity". Structure 14 (6): 983–92. June 2006. doi:10.1016/j.str.2006.04.001. PMID 16765891.
- ↑ 7.0 7.1 7.2 "Cooperation of breast cancer proteins PALB2 and piccolo BRCA2 in stimulating homologous recombination". Nature Structural & Molecular Biology 17 (10): 1247–54. October 2010. doi:10.1038/nsmb.1915. PMID 20871615.
- ↑ "Direct imaging of human Rad51 nucleoprotein dynamics on individual DNA molecules". Proceedings of the National Academy of Sciences of the United States of America 106 (2): 361–8. January 2009. doi:10.1073/pnas.0811965106. PMID 19122145.
- ↑ 9.0 9.1 9.2 "Over-expression of wild-type Rad51 correlates with histological grading of invasive ductal breast cancer". International Journal of Cancer 88 (6): 907–13. December 2000. doi:10.1002/1097-0215(20001215)88:6<907::aid-ijc11>3.0.co;2-4. PMID 11093813.
- ↑ 10.0 10.1 10.2 "RAD51 up-regulation bypasses BRCA1 function and is a common feature of BRCA1-deficient breast tumors". Cancer Research 67 (20): 9658–65. October 2007. doi:10.1158/0008-5472.CAN-07-0290. PMID 17942895.
- ↑ "RAD52 inactivation is synthetically lethal with deficiencies in BRCA1 and PALB2 in addition to BRCA2 through RAD51-mediated homologous recombination". Oncogene 32 (30): 3552–8. July 2013. doi:10.1038/onc.2012.391. PMID 22964643.
- ↑ "The consequences of Rad51 overexpression for normal and tumor cells". DNA Repair 7 (5): 686–93. May 2008. doi:10.1016/j.dnarep.2007.12.008. PMID 18243065.
- ↑ "High RAD51 mRNA expression characterize estrogen receptor-positive/progesteron receptor-negative breast cancer and is associated with patient's outcome". International Journal of Cancer 129 (3): 536–45. August 2011. doi:10.1002/ijc.25736. PMID 21064098.
- ↑ "Abnormal expression of BRCA1 and BRCA1-interactive DNA-repair proteins in breast carcinomas". International Journal of Cancer 88 (1): 28–36. October 2000. doi:10.1002/1097-0215(20001001)88:1<28::aid-ijc5>3.0.co;2-4. PMID 10962436.
- ↑ "Identification of differentially expressed genes in pancreatic cancer cells using cDNA microarray". Cancer Research 62 (10): 2890–6. May 2002. PMID 12019169.
- ↑ "DNA repair and recombination factor Rad51 is over-expressed in human pancreatic adenocarcinoma". Oncogene 19 (23): 2791–5. May 2000. doi:10.1038/sj.onc.1203578. PMID 10851081.
- ↑ "Pilot study examining tumor expression of RAD51 and clinical outcomes in human head cancers". International Journal of Oncology 28 (5): 1113–9. May 2006. doi:10.3892/ijo.28.5.1113. PMID 16596227.
- ↑ "Overexpression of RAD51 occurs in aggressive prostatic cancer". Histopathology 55 (6): 696–704. December 2009. doi:10.1111/j.1365-2559.2009.03448.x. PMID 20002770.
- ↑ "High-level expression of Rad51 is an independent prognostic marker of survival in non-small-cell lung cancer patients". British Journal of Cancer 93 (1): 137–43. July 2005. doi:10.1038/sj.bjc.6602665. PMID 15956972.
- ↑ "Rad51 overexpression contributes to chemoresistance in human soft tissue sarcoma cells: a role for p53/activator protein 2 transcriptional regulation". Molecular Cancer Therapeutics 6 (5): 1650–60. May 2007. doi:10.1158/1535-7163.MCT-06-0636. PMID 17513613.
- ↑ "Elevated expression of Rad51 is correlated with decreased survival in resectable esophageal squamous cell carcinoma". Journal of Surgical Oncology 104 (6): 617–22. November 2011. doi:10.1002/jso.22018. PMID 21744352.
- ↑ "Increased expression of SET domain-containing proteins and decreased expression of Rad51 in different classes of renal cell carcinoma". Bioscience Reports 36 (3): e00349. July 2016. doi:10.1042/BSR20160122. PMID 27170370.
- ↑ "Relative contribution of four nucleases, CtIP, Dna2, Exo1 and Mre11, to the initial step of DNA double-strand break repair by homologous recombination in both the chicken DT40 and human TK6 cell lines". Genes to Cells 20 (12): 1059–76. December 2015. doi:10.1111/gtc.12310. PMID 26525166.
- ↑ "BRCA1 and CtIP Are Both Required to Recruit Dna2 at Double-Strand Breaks in Homologous Recombination". PLOS ONE 10 (4): e0124495. 2015. doi:10.1371/journal.pone.0124495. PMID 25909997. Bibcode: 2015PLoSO..1024495H.
- ↑ "Domain mapping of the Rad51 paralog protein complexes". Nucleic Acids Research 32 (1): 169–78. 2004. doi:10.1093/nar/gkg925. PMID 14704354.
- ↑ 26.0 26.1 26.2 "Rad51 paralog complexes BCDX2 and CX3 act at different stages in the BRCA1-BRCA2-dependent homologous recombination pathway". Molecular and Cellular Biology 33 (2): 387–95. January 2013. doi:10.1128/MCB.00465-12. PMID 23149936.
- ↑ "Homologous recombination and human health: the roles of BRCA1, BRCA2, and associated proteins". Cold Spring Harbor Perspectives in Biology 7 (4): a016600. April 2015. doi:10.1101/cshperspect.a016600. PMID 25833843.
- ↑ 28.0 28.1 "Unraveling the mechanism of BRCA2 in homologous recombination". Nature Structural & Molecular Biology 18 (7): 748–54. July 2011. doi:10.1038/nsmb.2096. PMID 21731065.
- ↑ "Rad52 inactivation is synthetically lethal with BRCA2 deficiency". Proceedings of the National Academy of Sciences of the United States of America 108 (2): 686–91. January 2011. doi:10.1073/pnas.1010959107. PMID 21148102.
- ↑ "Meiotic Knockdown and Complementation Reveals Essential Role of RAD51 in Mouse Spermatogenesis". Cell Reports 18 (6): 1383–1394. February 2017. doi:10.1016/j.celrep.2017.01.024. PMID 28178517.
- ↑ "Spontaneous self-segregation of Rad51 and Dmc1 DNA recombinases within mixed recombinase filaments". The Journal of Biological Chemistry 293 (11): 4191–4200. March 2018. doi:10.1074/jbc.RA117.001143. PMID 29382724.
- ↑ "Enhancing survival of mouse oocytes following chemotherapy or aging by targeting Bax and Rad51". PLOS ONE 5 (2): e9204. February 2010. doi:10.1371/journal.pone.0009204. PMID 20169201. Bibcode: 2010PLoSO...5.9204K.
- ↑ "Most mammalian mRNAs are conserved targets of microRNAs". Genome Research 19 (1): 92–105. January 2009. doi:10.1101/gr.082701.108. PMID 18955434.
- ↑ "Specific activation of microRNA-127 with downregulation of the proto-oncogene BCL6 by chromatin-modifying drugs in human cancer cells". Cancer Cell 9 (6): 435–43. June 2006. doi:10.1016/j.ccr.2006.04.020. PMID 16766263.
- ↑ "Genetic unmasking of an epigenetically silenced microRNA in human cancer cells". Cancer Research 67 (4): 1424–9. February 2007. doi:10.1158/0008-5472.CAN-06-4218. PMID 17308079.
- ↑ 36.0 36.1 36.2 "Protective role of miR-155 in breast cancer through RAD51 targeting impairs homologous recombination after irradiation". Proceedings of the National Academy of Sciences of the United States of America 111 (12): 4536–41. March 2014. doi:10.1073/pnas.1402604111. PMID 24616504. Bibcode: 2014PNAS..111.4536G.
- ↑ 37.0 37.1 37.2 37.3 37.4 "MicroRNAs down-regulate homologous recombination in the G1 phase of cycling cells to maintain genomic stability". eLife 3: e02445. April 2014. doi:10.7554/eLife.02445. PMID 24843000.
- ↑ "miR-506: a regulator of chemo-sensitivity through suppression of the RAD51-homologous recombination axis". Chinese Journal of Cancer 34 (11): 485–7. September 2015. doi:10.1186/s40880-015-0049-z. PMID 26369335.
- ↑ "In Vivo Delivery of miR-34a Sensitizes Lung Tumors to Radiation Through RAD51 Regulation". Molecular Therapy: Nucleic Acids 4 (12): e270. December 2015. doi:10.1038/mtna.2015.47. PMID 26670277.
- ↑ 40.0 40.1 "Augmentation of response to chemotherapy by microRNA-506 through regulation of RAD51 in serous ovarian cancers". Journal of the National Cancer Institute 107 (7): djv108. July 2015. doi:10.1093/jnci/djv108. PMID 25995442.
- ↑ 41.0 41.1 "Inactivation of miR-34a by aberrant CpG methylation in multiple types of cancer". Cell Cycle 7 (16): 2591–600. August 2008. doi:10.4161/cc.7.16.6533. PMID 18719384.
- ↑ 42.0 42.1 "Development of a lung cancer therapeutic based on the tumor suppressor microRNA-34". Cancer Research 70 (14): 5923–30. July 2010. doi:10.1158/0008-5472.CAN-10-0655. PMID 20570894.
- ↑ 43.0 43.1 "Tumor-suppressive miR-34a induces senescence-like growth arrest through modulation of the E2F pathway in human colon cancer cells". Proceedings of the National Academy of Sciences of the United States of America 104 (39): 15472–7. September 2007. doi:10.1073/pnas.0707351104. PMID 17875987. Bibcode: 2007PNAS..10415472T.
- ↑ 44.0 44.1 "MicroRNA-34a functions as a potential tumor suppressor by inducing apoptosis in neuroblastoma cells". Oncogene 26 (34): 5017–22. July 2007. doi:10.1038/sj.onc.1210293. PMID 17297439.
- ↑ "MiR-96 downregulates REV1 and RAD51 to promote cellular sensitivity to cisplatin and PARP inhibition". Cancer Research 72 (16): 4037–46. August 2012. doi:10.1158/0008-5472.CAN-12-0103. PMID 22761336.
- ↑ "MicroRNA-203 Modulates the Radiation Sensitivity of Human Malignant Glioma Cells". International Journal of Radiation Oncology, Biology, Physics 94 (2): 412–20. February 2016. doi:10.1016/j.ijrobp.2015.10.001. PMID 26678661.
- ↑ "Systematic screen identifies miRNAs that target RAD51 and RAD51D to enhance chemosensitivity". Molecular Cancer Research 11 (12): 1564–73. December 2013. doi:10.1158/1541-7786.MCR-13-0292. PMID 24088786.
- ↑ "Highlight: BRCA1 and BRCA2 proteins in breast cancer". Microscopy Research and Technique 59 (1): 68–83. October 2002. doi:10.1002/jemt.10178. PMID 12242698.
- ↑ 49.0 49.1 "Insights into DNA recombination from the structure of a RAD51-BRCA2 complex". Nature 420 (6913): 287–93. November 2002. doi:10.1038/nature01230. PMID 12442171. Bibcode: 2002Natur.420..287P.
- ↑ "Molecular carcinogenesis of canine mammary tumors: news from an old disease". Veterinary Pathology 48 (1): 98–116. January 2011. doi:10.1177/0300985810390826. PMID 21149845.
- ↑ "Increased expression of BRCA2 and RAD51 in lymph node metastases of canine mammary adenocarcinomas". Veterinary Pathology 46 (3): 416–22. May 2009. doi:10.1354/vp.08-VP-0212-K-FL. PMID 19176491.
- ↑ "RAD51 protein expression is increased in canine mammary carcinomas". Veterinary Pathology 47 (1): 98–101. January 2010. doi:10.1177/0300985809353310. PMID 20080488.
- ↑ "The combined expression pattern of BMP2, LTBP4, and DERL1 discriminates malignant from benign canine mammary tumors". Veterinary Pathology 47 (3): 446–54. May 2010. doi:10.1177/0300985810363904. PMID 20375427.
- ↑ "A Dominant Mutation in Human RAD51 Reveals Its Function in DNA Interstrand Crosslink Repair Independent of Homologous Recombination". Molecular Cell 59 (3): 478–90. August 2015. doi:10.1016/j.molcel.2015.07.009. PMID 26253028.
- ↑ "A novel Fanconi anaemia subtype associated with a dominant-negative mutation in RAD51". Nature Communications 6: 8829. December 2015. doi:10.1038/ncomms9829. PMID 26681308. Bibcode: 2015NatCo...6.8829A.
- ↑ 56.0 56.1 56.2 "Radiation-induced assembly of Rad51 and Rad52 recombination complex requires ATM and c-Abl". The Journal of Biological Chemistry 274 (18): 12748–52. April 1999. doi:10.1074/jbc.274.18.12748. PMID 10212258.
- ↑ 57.0 57.1 57.2 57.3 57.4 57.5 "Regulation of BRCC, a holoenzyme complex containing BRCA1 and BRCA2, by a signalosome-like subunit and its role in DNA repair". Molecular Cell 12 (5): 1087–99. November 2003. doi:10.1016/s1097-2765(03)00424-6. PMID 14636569.
- ↑ 58.0 58.1 "Stable interaction between the products of the BRCA1 and BRCA2 tumor suppressor genes in mitotic and meiotic cells". Molecular Cell 2 (3): 317–28. September 1998. doi:10.1016/s1097-2765(00)80276-2. PMID 9774970.
- ↑ "Association of BRCA1 with Rad51 in mitotic and meiotic cells". Cell 88 (2): 265–75. January 1997. doi:10.1016/s0092-8674(00)81847-4. PMID 9008167.
- ↑ "Adenosine nucleotide modulates the physical interaction between hMSH2 and BRCA1". Oncogene 20 (34): 4640–9. August 2001. doi:10.1038/sj.onc.1204625. PMID 11498787.
- ↑ "Embryonic lethality and radiation hypersensitivity mediated by Rad51 in mice lacking Brca2". Nature 386 (6627): 804–10. April 1997. doi:10.1038/386804a0. PMID 9126738. Bibcode: 1997Natur.386..804S.
- ↑ "M phase-specific phosphorylation of BRCA2 by Polo-like kinase 1 correlates with the dissociation of the BRCA2-P/CAF complex". The Journal of Biological Chemistry 278 (38): 35979–87. September 2003. doi:10.1074/jbc.M210659200. PMID 12815053.
- ↑ "Dynamic control of Rad51 recombinase by self-association and interaction with BRCA2". Molecular Cell 12 (4): 1029–41. October 2003. doi:10.1016/s1097-2765(03)00394-0. PMID 14580352.
- ↑ "The BRC repeats in BRCA2 are critical for RAD51 binding and resistance to methyl methanesulfonate treatment". Proceedings of the National Academy of Sciences of the United States of America 95 (9): 5287–92. April 1998. doi:10.1073/pnas.95.9.5287. PMID 9560268. Bibcode: 1998PNAS...95.5287C.
- ↑ "Analysis of murine Brca2 reveals conservation of protein-protein interactions but differences in nuclear localization signals". The Journal of Biological Chemistry 276 (40): 37640–8. October 2001. doi:10.1074/jbc.M106281200. PMID 11477095.
- ↑ "RAD51 interacts with the evolutionarily conserved BRC motifs in the human breast cancer susceptibility gene brca2". The Journal of Biological Chemistry 272 (51): 31941–4. December 1997. doi:10.1074/jbc.272.51.31941. PMID 9405383.
- ↑ "Multiple possible sites of BRCA2 interacting with DNA repair protein RAD51". Genes, Chromosomes & Cancer 21 (3): 217–22. March 1998. doi:10.1002/(SICI)1098-2264(199803)21:3<217::AID-GCC5>3.0.CO;2-2. PMID 9523196.
- ↑ "RAD51 localization and activation following DNA damage". Philosophical Transactions of the Royal Society of London. Series B, Biological Sciences 359 (1441): 87–93. January 2004. doi:10.1098/rstb.2003.1368. PMID 15065660.
- ↑ "Inhibition of breast and brain cancer cell growth by BCCIPalpha, an evolutionarily conserved nuclear protein that interacts with BRCA2". Oncogene 20 (3): 336–45. January 2001. doi:10.1038/sj.onc.1204098. PMID 11313963.
- ↑ "The BRCA2 gene product functionally interacts with p53 and RAD51". Proceedings of the National Academy of Sciences of the United States of America 95 (23): 13869–74. November 1998. doi:10.1073/pnas.95.23.13869. PMID 9811893. Bibcode: 1998PNAS...9513869M.
- ↑ "Potential role for the BLM helicase in recombinational repair via a conserved interaction with RAD51". The Journal of Biological Chemistry 276 (22): 19375–81. June 2001. doi:10.1074/jbc.M009471200. PMID 11278509.
- ↑ "The meiosis-specific recombinase hDmc1 forms ring structures and interacts with hRad51". The EMBO Journal 18 (22): 6552–60. November 1999. doi:10.1093/emboj/18.22.6552. PMID 10562567.
- ↑ "Homologous DNA pairing by human recombination factors Rad51 and Rad54". The Journal of Biological Chemistry 277 (45): 42790–4. November 2002. doi:10.1074/jbc.M208004200. PMID 12205100.
- ↑ "p53 is linked directly to homologous recombination processes via RAD51/RecA protein interaction". The EMBO Journal 15 (8): 1992–2002. April 1996. doi:10.1002/j.1460-2075.1996.tb00550.x. PMID 8617246.
- ↑ "Interaction of p53 with the human Rad51 protein". Nucleic Acids Research 25 (19): 3868–74. October 1997. doi:10.1093/nar/25.19.3868. PMID 9380510.
- ↑ "A novel human rad54 homologue, Rad54B, associates with Rad51". The Journal of Biological Chemistry 275 (34): 26316–21. August 2000. doi:10.1074/jbc.M910306199. PMID 10851248.
- ↑ "Mammalian ubiquitin-conjugating enzyme Ubc9 interacts with Rad51 recombination protein and localizes in synaptonemal complexes". Proceedings of the National Academy of Sciences of the United States of America 93 (7): 2958–63. April 1996. doi:10.1073/pnas.93.7.2958. PMID 8610150. Bibcode: 1996PNAS...93.2958K.
- ↑ "Associations of UBE2I with RAD52, UBL1, p53, and RAD51 proteins in a yeast two-hybrid system". Genomics 37 (2): 183–6. October 1996. doi:10.1006/geno.1996.0540. PMID 8921390. https://zenodo.org/record/1229705.
External links
- RAD51+Protein at the US National Library of Medicine Medical Subject Headings (MeSH)
 |