Biology:Collagen helix
| Collagen triple helix | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
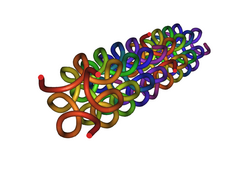 Model of a collagen helix.[1] | |||||||||||
| Identifiers | |||||||||||
| Symbol | Collagen | ||||||||||
| Pfam | PF01391 | ||||||||||
| InterPro | IPR008160 | ||||||||||
| SCOP2 | 1a9a / SCOPe / SUPFAM | ||||||||||
| |||||||||||

In molecular biology, the collagen triple helix or type-2 helix is the main secondary structure of various types of fibrous collagen, including type I collagen. In 1954, Ramachandran & Kartha (13, 14) advanced a structure for the collagen triple helix on the basis of fiber diffraction data. It consists of a triple helix made of the repetitious amino acid sequence glycine-X-Y, where X and Y are frequently proline or hydroxyproline.[2][3] Collagen folded into a triple helix is known as tropocollagen. Collagen triple helices are often bundled into fibrils which themselves form larger fibres, as in tendons.
Structure
Glycine, proline, and hydroxyproline must be in their designated positions with the correct configuration. For example, hydroxyproline in the Y position increases the thermal stability of the triple helix, but not when it is located in the X position.[4] The thermal stabilization is also hindered when the hydroxyl group has the wrong configuration. Due to the high abundance of glycine and proline contents, collagen fails to form a regular α-helix and β-sheet structure. Three left-handed helical strands twist to form a right-handed triple helix.[5] A collagen triple helix has 3.3 residues per turn.[6]
Each of the three chains is stabilized by the steric repulsion due to the pyrrolidine rings of proline and hydroxyproline residues. The pyrrolidine rings keep out of each other's way when the polypeptide chain assumes this extended helical form, which is much more open than the tightly coiled form of the alpha helix. The three chains are hydrogen bonded to each other. The hydrogen bond donors are the peptide NH groups of glycine residues. The hydrogen bond acceptors are the CO groups of residues on the other chains. The OH group of hydroxyproline does not participate in hydrogen bonding but stabilises the trans isomer of proline by stereoelectronic effects, therefore stabilizing the entire triple helix.
The rise of the collagen helix (superhelix) is 2.9 Å (0.29 nm) per residue. The center of the collagen triple helix is very small and hydrophobic, and every third residue of the helix must have contact with the center.[7] Due to the very tiny and tight space at the center, only the small hydrogen of the glycine side chain is capable of interacting with the center.[7] This contact is impossible even when a slightly bigger amino acid residue is present other than glycine.
References
- ↑ "Crystal structure of the collagen triple helix model [(Pro-Pro-Gly)(10)(3)"]. Protein Sci. 11 (2): 262–70. February 2002. doi:10.1110/ps.32602. PMID 11790836.
- ↑ "Collagen structure: the Madras triple helix and the current scenario". IUBMB Life 57 (3): 161–72. March 2005. doi:10.1080/15216540500090710. PMID 16036578.
- ↑ Saad, Mohamed (Oct 1994). Low resolution structure and packing investigations of collagen crystalline domains in tendon using Synchrotron Radiation X-rays, Structure factors determination, evaluation of Isomorphous Replacement methods and other modeling.. PhD Thesis, Université Joseph Fourier Grenoble I. pp. 1–221. doi:10.13140/2.1.4776.7844. https://drive.google.com/open?id=0B3L_EN9hIuFTTkhuN2lrWEU4RDQ&authuser=0.
- ↑ Berisio R, Vitagliano L, Mazzarella L, Zagari A (February 2002). "Crystal structure of the collagen triple helix model [(Pro-Pro-Gly)(10)](3)". Protein Sci. 11 (2): 262–70. doi:10.1110/ps.32602. PMC 2373432. PMID 11790836.
- ↑ Bella, Jordi. “Collagen Structure: New Tricks from a Very Old Dog.” The Biochemical Journal, vol. 473, no. 8, 2016, pp. 1001–1025.
- ↑ Harpers Illustrated Biochemistry, 30th edition
- ↑ 7.0 7.1 Brodsky, Barbara, et al. “Triple‐Helical Peptides: An Approach to Collagen Conformation, Stability, and Self‐Association.” Biopolymers, vol. 89, no. 5, 2008, pp. 345–353.
 |