Biology:Zaire ebolavirus
| Zaire ebolavirus | |
|---|---|
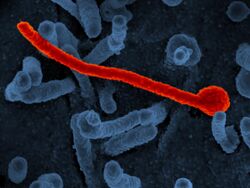
| File:Ebola Virus - Electron Micrograph.tiff | |
| Colorized scanning electron micrograph of Ebola virus particles (green) found both as extracellular particles and budding particles from a chronically infected African Green Monkey Kidney cell (blue); 20,000x magnification | |
| Virus classification | |
| Script error: No such module "Taxobox ranks".: | Virus |
| Script error: No such module "Taxobox ranks".: | Riboviria |
| Script error: No such module "Taxobox ranks".: | Orthornavirae |
| Script error: No such module "Taxobox ranks".: | Negarnaviricota |
| Script error: No such module "Taxobox ranks".: | Monjiviricetes |
| Script error: No such module "Taxobox ranks".: | Mononegavirales |
| Script error: No such module "Taxobox ranks".: | Filoviridae |
| Script error: No such module "Taxobox ranks".: | Ebolavirus |
| Script error: No such module "Taxobox ranks".: | <div style="display:inline" class="script error: no such module "taxobox ranks".">Zaire ebolavirus |
Zaire ebolavirus, more commonly known as Ebola virus (/iˈboʊlə, ɪ-/; EBOV), is one of six known species within the genus Ebolavirus.[1] Four of the six known ebolaviruses, including EBOV, cause a severe and often fatal hemorrhagic fever in humans and other mammals, known as Ebola virus disease (EVD). Ebola virus has caused the majority of human deaths from EVD, and was the cause of the 2013–2016 epidemic in western Africa,[2] which resulted in at least {{#section:Ebola virus epidemic in West Africa|cases}} suspected cases and {{#section:Ebola virus epidemic in West Africa|deaths}} confirmed deaths.{{#section:Ebola virus epidemic in West Africa|caserefs}}[3]
Ebola virus and its genus were both originally named for Zaire (now the Democratic Republic of the Congo), the country where it was first described,[1] and was at first suspected to be a new "strain" of the closely related Marburg virus.[4][5] The virus was renamed "Ebola virus" in 2010 to avoid confusion. Ebola virus is the single member of the species Zaire ebolavirus, which is assigned to the genus Ebolavirus, family Filoviridae, order Mononegavirales. The members of the species are called Zaire ebolaviruses.[1][6] The natural reservoir of Ebola virus is believed to be bats, particularly fruit bats,[7] and it is primarily transmitted between humans and from animals to humans through body fluids.[8]
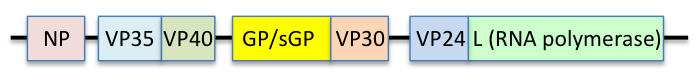
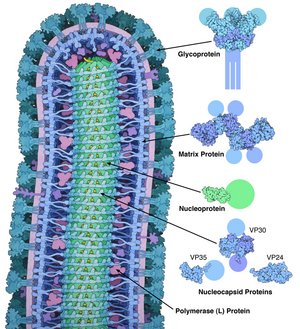
The EBOV genome is a single-stranded RNA, approximately 19,000 nucleotides long. It encodes seven structural proteins: nucleoprotein (NP), polymerase cofactor (VP35), (VP40), GP, transcription activator (VP30), VP24, and RNA-dependent RNA polymerase (L).[9]
Because of its high fatality rate (up to 83 to 90 percent),[10][11] EBOV is also listed as a select agent, World Health Organization Risk Group 4 Pathogen (requiring Biosafety Level 4-equivalent containment), a US National Institutes of Health/National Institute of Allergy and Infectious Diseases Category A Priority Pathogen, US CDC Centers for Disease Control and Prevention Category A Bioterrorism Agent, and a Biological Agent for Export Control by the Australia Group.[citation needed]
Structure
EBOV carries a negative-sense RNA genome in virions that are cylindrical/tubular, and contain viral envelope, matrix, and nucleocapsid components. The overall cylinders are generally approximately 80 nm in diameter, and have a virally encoded glycoprotein (GP) projecting as 7–10 nm long spikes from its lipid bilayer surface.[12] The cylinders are of variable length, typically 800 nm, but sometimes up to 1000 nm long. The outer viral envelope of the virion is derived by budding from domains of host cell membrane into which the GP spikes have been inserted during their biosynthesis. Individual GP molecules appear with spacings of about 10 nm. Viral proteins VP40 and VP24 are located between the envelope and the nucleocapsid (see following), in the matrix space.[13] At the center of the virion structure is the nucleocapsid, which is composed of a series of viral proteins attached to an 18–19 kb linear, negative-sense RNA without 3′-polyadenylation or 5′-capping (see following); the RNA is helically wound and complexed with the NP, VP35, VP30, and L proteins; this helix has a diameter of 80 nm.[14][15][16]
The overall shape of the virions after purification and visualization (e.g., by ultracentrifugation and electron microscopy, respectively) varies considerably; simple cylinders are far less prevalent than structures showing reversed direction, branches, and loops (e.g., U-, shepherd's crook-, 9-, or eye bolt-shapes, or other or circular/coiled appearances), the origin of which may be in the laboratory techniques applied.[17][18] The characteristic "threadlike" structure is, however, a more general morphologic characteristic of filoviruses (alongside their GP-decorated viral envelope, RNA nucleocapsid, etc.).[17]
Genome
Each virion contains one molecule of linear, single-stranded, negative-sense RNA, 18,959 to 18,961 nucleotides in length.[19] The 3′ terminus is not polyadenylated and the 5′ end is not capped. This viral genome codes for seven structural proteins and one non-structural protein. The gene order is 3′ – leader – NP – VP35 – VP40 – GP/sGP – VP30 – VP24 – L – trailer – 5′; with the leader and trailer being non-transcribed regions, which carry important signals to control transcription, replication, and packaging of the viral genomes into new virions. Sections of the NP, VP35 and the L genes from filoviruses have been identified as endogenous in the genomes of several groups of small mammals.[20][21][22]
It was found that 472 nucleotides from the 3' end and 731 nucleotides from the 5' end are sufficient for replication of a viral "minigenome", though not sufficient for infection.[17] Virus sequencing from 78 patients with confirmed Ebola virus disease, representing more than 70% of cases diagnosed in Sierra Leone from late May to mid-June 2014,[23][24] provided evidence that the 2014 outbreak was no longer being fed by new contacts with its natural reservoir. Using third-generation sequencing technology, investigators were able to sequence samples as quickly as 48 hours.[25] Like other RNA viruses,[23] Ebola virus mutates rapidly, both within a person during the progression of disease and in the reservoir among the local human population.[24] The observed mutation rate of 2.0 x 10−3 substitutions per site per year is as fast as that of seasonal influenza.[26]
| Symbol | Name | UniProt | Function |
|---|---|---|---|
| NP | Nucleoprotein | P18272 | Wraps genome for protection from nucleases and innate immunity. |
| VP35 | Polymerase cofactor VP35 | Q05127 | Polymerase cofactor; suppresses innate immunity by binding RNA. |
| VP40 | Matrix protein VP40 | Q05128 | Matrix. |
| GP | Envelope glycoprotein | Q05320 | Cleaved by host furin into GP1/2 to form envelope with spikes. Also makes shed GP as a decoy. |
| sGP | Pre-small/secreted glycoprotein | P60170 | Shares ORF with GP. Cleaved by host furin into sGP (anti-inflammatory) and delta-peptide (viroporin). |
| ssGP | Super small secreted glycoprotein | Q9YMG2 | Shares ORF with GP; created by mRNA editing. Unknown function. |
| VP30 | Hexameric zinc-finger protein VP30 | Q05323 | Transcriptional activator. |
| VP24 | Membrane-associated protein VP24 | Q05322 | Blocks IFN-alpha/beta and IFN-gamma signaling. |
| L | RNA-directed RNA polymerase L | Q05318 | RNA replicase. |
Entry
There are two candidates for host cell entry proteins. The first is a cholesterol transporter protein, the host-encoded Niemann–Pick C1 (NPC1), which appears to be essential for entry of Ebola virions into the host cell and for its ultimate replication.[27][28] In one study, mice with one copy of the NPC1 gene removed showed an 80 percent survival rate fifteen days after exposure to mouse-adapted Ebola virus, while only 10 percent of unmodified mice survived this long.[27] In another study, small molecules were shown to inhibit Ebola virus infection by preventing viral envelope glycoprotein (GP) from binding to NPC1.[28][29] Hence, NPC1 was shown to be critical to entry of this filovirus, because it mediates infection by binding directly to viral GP.[28]
When cells from Niemann–Pick Type C individuals lacking this transporter were exposed to Ebola virus in the laboratory, the cells survived and appeared impervious to the virus, further indicating that Ebola relies on NPC1 to enter cells;[27] mutations in the NPC1 gene in humans were conjectured as a possible mode to make some individuals resistant to this deadly viral disease. The same studies described similar results regarding NPC1's role in virus entry for Marburg virus, a related filovirus.[27] A further study has also presented evidence that NPC1 is the critical receptor mediating Ebola infection via its direct binding to the viral GP, and that it is the second "lysosomal" domain of NPC1 that mediates this binding.[30]
The second candidate is TIM-1 (a.k.a. HAVCR1).[31] TIM-1 was shown to bind to the receptor binding domain of the EBOV glycoprotein, to increase the receptivity of Vero cells. Silencing its effect with siRNA prevented infection of Vero cells. TIM1 is expressed in tissues known to be seriously impacted by EBOV lysis (trachea, cornea, and conjunctiva). A monoclonal antibody against the IgV domain of TIM-1, ARD5, blocked EBOV binding and infection. Together, these studies suggest NPC1 and TIM-1 may be potential therapeutic targets for an Ebola anti-viral drug and as a basis for a rapid field diagnostic assay.[citation needed]
Replication
Being acellular, viruses such as Ebola do not replicate through any type of cell division; rather, they use a combination of host- and virally encoded enzymes, alongside host cell structures, to produce multiple copies of themselves. These then self-assemble into viral macromolecular structures in the host cell.[32] The virus completes a set of steps when infecting each individual cell. The virus begins its attack by attaching to host receptors through the glycoprotein (GP) surface peplomer and is endocytosed into macropinosomes in the host cell.[33] To penetrate the cell, the viral membrane fuses with vesicle membrane, and the nucleocapsid is released into the cytoplasm. Encapsidated, negative-sense genomic ssRNA is used as a template for the synthesis (3'–5') of polyadenylated, monocistronic mRNAs and, using the host cell's ribosomes, tRNA molecules, etc., the mRNA is translated into individual viral proteins.[34][35][36]
These viral proteins are processed: a glycoprotein precursor (GP0) is cleaved to GP1 and GP2, which are then heavily glycosylated using cellular enzymes and substrates. These two molecules assemble, first into heterodimers, and then into trimers to give the surface peplomers. Secreted glycoprotein (sGP) precursor is cleaved to sGP and delta peptide, both of which are released from the cell. As viral protein levels rise, a switch occurs from translation to replication. Using the negative-sense genomic RNA as a template, a complementary +ssRNA is synthesized; this is then used as a template for the synthesis of new genomic (-)ssRNA, which is rapidly encapsidated. The newly formed nucleocapsids and envelope proteins associate at the host cell's plasma membrane; budding occurs, destroying the cell.[citation needed]
Ecology
Ebola virus is a zoonotic pathogen. Intermediary hosts have been reported to be "various species of fruit bats ... throughout central and sub-Saharan Africa". Evidence of infection in bats has been detected through molecular and serologic means. However, ebolaviruses have not been isolated in bats.[7][37] End hosts are humans and great apes, infected through bat contact or through other end hosts. Pigs in the Philippines have been reported to be infected with Reston virus, so other interim or amplifying hosts may exist.[37] Ebola virus outbreaks tend to occur when temperatures are lower and humidity is higher than usual for Africa.[38] Even after a person recovers from the acute phase of the disease, Ebola virus survives for months in certain organs such as the eyes and testes.[39]
Ebola virus disease
Zaire ebolavirus is one of the four ebolaviruses known to cause disease in humans. It has the highest case-fatality rate of these ebolaviruses, averaging 83 percent since the first outbreaks in 1976, although a fatality rate of up to 90 percent was recorded in one outbreak in the Republic of the Congo between December 2002 and April 2003. There have also been more outbreaks of Zaire ebolavirus than of any other ebolavirus. The first outbreak occurred on 26 August 1976 in Yambuku.[40] The first recorded case was Mabalo Lokela, a 44‑year-old schoolteacher. The symptoms resembled malaria, and subsequent patients received quinine. Transmission has been attributed to reuse of unsterilized needles and close personal contact, body fluids and places where the person has touched. During the 1976 Ebola outbreak in Zaire, Ngoy Mushola travelled from Bumba to Yambuku, where he recorded the first clinical description of the disease in his daily log:
The illness is characterized with a high temperature of about 39°C, hematemesis, diarrhea with blood, retrosternal abdominal pain, prostration with "heavy" articulations, and rapid evolution death after a mean of three days.[41]
Since the first recorded clinical description of the disease during 1976 in Zaire, the recent Ebola outbreak that started in March 2014, in addition, reached epidemic proportions and has killed more than 8000 people as of January 2015. This outbreak was centered in West Africa, an area that had not previously been affected by the disease. The toll was particularly grave in three countries: Guinea, Liberia, and Sierra Leone. A few cases were also reported in countries outside of West Africa, all related to international travelers who were exposed in the most affected regions and later showed symptoms of Ebola fever after reaching their destinations.[42]
The severity of the disease in humans varies widely, from rapid fatality to mild illness or even asymptomatic response.[43] Studies of outbreaks in the late twentieth century failed to find a correlation between the disease severity and the genetic nature of the virus. Hence the variability in the severity of illness was suspected to correlate with genetic differences in the victims. This has been difficult to study in animal models that respond to the virus with hemorrhagic fever in a similar manner as humans, because typical mouse models do not so respond, and the required large numbers of appropriate test subjects are not easily available. In late October 2014, a publication reported a study of the response to a mouse-adapted strain of Zaire ebolavirus presented by a genetically diverse population of mice that was bred to have a range of responses to the virus that includes fatality from hemorrhagic fever.[44]
Vaccine
In December 2016, a study found the VSV-EBOV vaccine to be 70–100% effective against the Zaire ebola virus (not the Sudan ebolavirus), making it the first vaccine against the disease.[45][46] VSV-EBOV was approved by the U.S. Food and Drug Administration in December 2019.[47]
History and nomenclature
Ebola virus was first identified as a possible new "strain" of Marburg virus in 1976.[4][5][48] The International Committee on Taxonomy of Viruses (ICTV) identifies Ebola virus as species Zaire ebolavirus, which is part of the genus Ebolavirus, family Filoviridae, order Mononegavirales. The name "Ebola virus" is derived from the Ebola River—a river that was at first thought to be in close proximity to the area in Democratic Republic of Congo, previously called Zaire, where the 1976 Zaire Ebola virus outbreak occurred—and the taxonomic suffix virus.[1][4][5][49]
In 1998, the virus name was changed to "Zaire Ebola virus"[50][51] and in 2002 to species Zaire ebolavirus.[52][53] However, most scientific articles continued to refer to "Ebola virus" or used the terms "Ebola virus" and "Zaire ebolavirus" in parallel. Consequently, in 2010, a group of researchers recommended that the name "Ebola virus" be adopted for a subclassification within the species Zaire ebolavirus, with the corresponding abbreviation EBOV.[1] Previous abbreviations for the virus were EBOV-Z (for "Ebola virus Zaire") and ZEBOV (for "Zaire Ebola virus" or "Zaire ebolavirus"). In 2011, the ICTV explicitly rejected a proposal (2010.010bV) to recognize this name, as ICTV does not designate names for subtypes, variants, strains, or other subspecies level groupings.[54] At present, ICTV does not officially recognize "Ebola virus" as a taxonomic rank, but rather continues to use and recommend only the species designation Zaire ebolavirus.[55] The prototype Ebola virus, variant Mayinga (EBOV/May), was named for Mayinga N'Seka, a nurse who died during the 1976 Zaire outbreak.[1][56][57]
The name Zaire ebolavirus is derived from Zaire and the taxonomic suffix ebolavirus (which denotes an ebolavirus species and refers to the Ebola River).[1] According to the rules for taxon naming established by the International Committee on Taxonomy of Viruses (ICTV), the name Zaire ebolavirus is always to be capitalized, italicized, and to be preceded by the word "species". The names of its members (Zaire ebolaviruses) are to be capitalized, are not italicized, and used without articles.[1]
Virus inclusion criteria
A virus of the genus Ebolavirus is a member of the species Zaire ebolavirus if:[1]
- it is endemic in the Democratic Republic of the Congo, Gabon, or the Republic of the Congo
- it has a genome with two or three gene overlaps (VP35/VP40, GP/VP30, VP24/L)
- it has a genomic sequence that differs from the type virus EBOV/May by less than 30%
Evolution
Zaire ebolavirus diverged from its ancestors between 1960 and 1976.[58] The genetic diversity of Ebolavirus remained constant before 1900.[58][59] Then, around the 1960s, most likely due to climate change or human activities, the genetic diversity of the virus dropped rapidly and most lineages became extinct.[59] As the number of susceptible hosts declines, so does the effective population size and its genetic diversity. This genetic bottleneck effect has implications for the species' ability to cause Ebola virus disease in human hosts.[citation needed]
A recombination event between Zaire ebolavirus lineages likely took place between 1996 and 2001 in wild apes giving rise to recombinant progeny viruses.[60] These recombinant viruses appear to have been responsible for a series of outbreaks among humans in Central Africa in 2001–2003.[60]
Zaire ebolavirus – Makona variant caused the 2014 West Africa outbreak.[61] The outbreak was characterized by the longest instance of human-to-human transmission of the viral species.[61] Pressures to adapt to the human host were seen at this time, however, no phenotypic changes in the virus (such as increased transmission, increased immune evasion by the virus) were seen.[citation needed]
In literature
- Alex Kava's 2008 crime novel, Exposed, focuses on the virus as a serial killer's weapon of choice.[62]
- William Close's 1995 Ebola: A Documentary Novel of Its First Explosion and 2002 Ebola: Through the Eyes of the People focused on individuals' reactions to the 1976 Ebola outbreak in Zaire.[63][64][65][66]
- The Hot Zone: A Terrifying True Story: A 1994 best-selling book by Richard Preston about Ebola virus and related viruses, including an account of the outbreak of an Ebolavirus in primates housed in a quarantine facility in Reston, Virginia, USA[67]
- Tom Clancy's 1996 novel, Executive Orders, involves a Middle Eastern terrorist attack on the United States using an airborne form of a deadly Ebola virus named "Ebola Mayinga".[68][69]
References
- ↑ 1.0 1.1 1.2 1.3 1.4 1.5 1.6 1.7 1.8 "Proposal for a revised taxonomy of the family Filoviridae: Classification, names of taxa and viruses, and virus abbreviations". Archives of Virology 155 (12): 2083–2103. 2010. doi:10.1007/s00705-010-0814-x. PMID 21046175.
- ↑ Na, Woonsung; Park, Nanuri; Yeom, Minju; Song, Daesub (4 December 2016). "Ebola outbreak in Western Africa 2014: what is going on with Ebola virus?". Clinical and Experimental Vaccine Research 4 (1): 17–22. doi:10.7774/cevr.2015.4.1.17. ISSN 2287-3651. PMID 25648530.
- ↑ "Ebola virus disease outbreak". https://www.who.int/csr/disease/ebola/en/.
- ↑ 4.0 4.1 4.2 "Isolation of Marburg-like virus from a case of haemorrhagic fever in Zaire". Lancet 309 (8011): 573–574. 1977. doi:10.1016/s0140-6736(77)92002-5. PMID 65663.
- ↑ 5.0 5.1 5.2 "Viral haemorrhagic fever in southern Sudan and northern Zaire. Preliminary studies on the aetiological agent". Lancet 309 (8011): 571–573. 1977. doi:10.1016/s0140-6736(77)92001-3. PMID 65662.
- ↑ WHO. "Ebola virus disease". https://www.who.int/mediacentre/factsheets/fs103/en/.
- ↑ 7.0 7.1 Quammen, David (2014-12-30). "Insect-Eating Bat May Be Origin of Ebola Outbreak, New Study Suggests". Washington, DC: National Geographic Society. http://news.nationalgeographic.com/news/2014/12/141230-ebola-virus-origin-insect-bats-meliandou-reservoir-host/.
- ↑ Angier, Natalie (October 27, 2014). "Killers in a Cell but on the Loose – Ebola and the Vast Viral Universe". New York Times. https://www.nytimes.com/2014/10/28/science/ebola-and-the-vast-viral-universe.html.
- ↑ Nanbo, Asuka; Watanabe, Shinji; Halfmann, Peter; Kawaoka, Yoshihiro (4 Feb 2013). "The spatio-temporal distribution dynamics of Ebola virus proteins and RNA in infected cells". Scientific Reports 3: 1206. doi:10.1038/srep01206. PMID 23383374. Bibcode: 2013NatSR...3E1206N.
- ↑ "Ebola virus disease Fact sheet N°103". March 2014. https://www.who.int/mediacentre/factsheets/fs103/en/.
- ↑ Virus Taxonomy – Eighth Report of the International Committee on Taxonomy of Viruses. Oxford: Elsevier/Academic Press. 2005. p. 648. ISBN 978-0080575483. https://books.google.com/books?id=9Wy7Jgy5RWYC.
- ↑ Klenk, H.-D.; Feldmann, H., eds (2004). Ebola and Marburg Viruses – Molecular and Cellular Biology. Wymondham, Norfolk, UK: Horizon Bioscience. p. 28. ISBN 978-0-9545232-3-7.
- ↑ Feldmann, H. K. (1993). "Molecular biology and evolution of filoviruses". Unconventional Agents and Unclassified Viruses. Archives of Virology. 7. 81–100. doi:10.1007/978-3-7091-9300-6_8. ISBN 978-3211824801.
- ↑ Lee, Jeffrey E; Saphire, Erica Ollmann (2009). "Ebolavirus glycoprotein structure and mechanism of entry". Future Virology 4 (6): 621–635. doi:10.2217/fvl.09.56. ISSN 1746-0794. PMID 20198110.
- ↑ "Molecular mechanisms of Ebola virus pathogenesis: focus on cell death". Cell Death and Differentiation 22 (8): 1250–1259. 4 December 2016. doi:10.1038/cdd.2015.67. ISSN 1350-9047. PMID 26024394.
- ↑ Swetha, Rayapadi G.; Ramaiah, Sudha; Anbarasu, Anand; Sekar, Kanagaraj (2016). "Ebolavirus Database: Gene and Protein Information Resource for Ebolaviruses". Advances in Bioinformatics 2016: 1673284. doi:10.1155/2016/1673284. ISSN 1687-8027. PMID 27190508.
- ↑ 17.0 17.1 17.2 Klenk, H.-D.; Feldmann, H., eds (2004). Ebola and Marburg Viruses: Molecular and Cellular Biology. Horizon Bioscience. ISBN 978-1904933496.[page needed]
- ↑ Hillman, H. (1991). The Case for New Paradigms in Cell Biology and in Neurobiology. Edwin Mellen Press.
- ↑ Zaire ebolavirus isolate H.sapiens-wt/GIN/2014/Makona-Kissidougou-C15, complete genome, GenBank
- ↑ "Filoviruses are ancient and integrated into mammalian genomes". BMC Evolutionary Biology 10 (1): 193. 2010. doi:10.1186/1471-2148-10-193. PMID 20569424. Bibcode: 2010BMCEE..10..193T.
- ↑ Belyi, V. A.; Levine, A. J.; Skalka, A. M. (2010). Buchmeier, Michael J.. ed. "Unexpected Inheritance: Multiple Integrations of Ancient Bornavirus and Ebolavirus/Marburgvirus Sequences in Vertebrate Genomes". PLOS Pathogens 6 (7): e1001030. doi:10.1371/journal.ppat.1001030. PMID 20686665.
- ↑ "Evidence that ebolaviruses and cuevaviruses have been diverging from marburgviruses since the Miocene". PeerJ 2: e556. 2014. doi:10.7717/peerj.556. PMID 25237605.
- ↑ 23.0 23.1 Richard Preston (27 October 2014). "The Ebola Wars". The New Yorker (New York: Condé Nast). https://www.newyorker.com/magazine/2014/10/27/ebola-wars. Retrieved 20 October 2014.
- ↑ 24.0 24.1 Gire, Stephen K. (2014). "Genomic surveillance elucidates Ebola virus origin and transmission during the 2014 outbreak". Science 345 (6202): 1369–1372. doi:10.1126/science.1259657. PMID 25214632. Bibcode: 2014Sci...345.1369G.
- ↑ Check Hayden, Erika (2015-05-05). "Pint-sized DNA sequencer impresses first users" (in en). Nature 521 (7550): 15–16. doi:10.1038/521015a. ISSN 0028-0836. PMID 25951262. Bibcode: 2015Natur.521...15C.
- ↑ "Rates of molecular evolution in RNA viruses: A quantitative phylogenetic analysis". Journal of Molecular Evolution 54 (2): 156–165. 2002. doi:10.1007/s00239-001-0064-3. PMID 11821909. Bibcode: 2002JMolE..54..156J.
- ↑ 27.0 27.1 27.2 27.3 "Ebola virus entry requires the cholesterol transporter Niemann-Pick C1". Nature 477 (7364): 340–343. September 2011. doi:10.1038/nature10348. PMID 21866103. Bibcode: 2011Natur.477..340C.
- Amanda Schaffer (January 16, 2012). "Key Protein May Give Ebola Virus Its Opening". The New York Times. https://www.nytimes.com/2012/01/17/health/npc1-protein-may-give-ebola-its-opening.html.
- ↑ 28.0 28.1 28.2 "Small molecule inhibitors reveal Niemann-Pick C1 is essential for Ebola virus infection". Nature 477 (7364): 344–348. September 2011. doi:10.1038/nature10380. PMID 21866101. Bibcode: 2011Natur.477..344C.
- Amanda Schaffer (January 16, 2012). "Key Protein May Give Ebola Virus Its Opening". The New York Times. https://www.nytimes.com/2012/01/17/health/npc1-protein-may-give-ebola-its-opening.html.
- ↑ "Achilles heel of Ebola viral entry". Nat Rev Drug Discov 10 (10): 731. October 2011. doi:10.1038/nrd3568. PMID 21959282.
- ↑ "Ebola virus entry requires the host-programmed recognition of an intracellular receptor". EMBO Journal 31 (8): 1947–1960. March 2012. doi:10.1038/emboj.2012.53. PMID 22395071.
- ↑ "T-cell immunoglobulin and mucin domain 1 (TIM-1) is a receptor for Zaire Ebolavirus and Lake Victoria Marburgvirus". Proceedings of the National Academy of Sciences of the United States of America 108 (20): 8426–8431. May 2011. doi:10.1073/pnas.1019030108. PMID 21536871. Bibcode: 2011PNAS..108.8426K.
- ↑ Biomarker Database. Ebola virus. Korea National Institute of Health. http://biomarker.cdc.go.kr:8080/pathogen/pathogen_view_en.jsp?pclass=2&id=44. Retrieved 2009-05-31.
- ↑ Basler, Christopher F., ed (2010). "Cellular Entry of Ebola Virus Involves Uptake by a Macropinocytosis-Like Mechanism and Subsequent Trafficking through Early and Late Endosomes". PLOS Pathogens 6 (9): e1001110. doi:10.1371/journal.ppat.1001110. PMID 20862315.
- ↑ Mühlberger, Elke (4 December 2016). "Filovirus replication and transcription". Future Virology 2 (2): 205–215. doi:10.2217/17460794.2.2.205. ISSN 1746-0794. PMID 24093048.
- ↑ Feldmann, H.; Klenk, H.-D. (1996). Filoviruses. University of Texas Medical Branch at Galveston. ISBN 978-0963117212. https://www.ncbi.nlm.nih.gov/books/NBK8129/. Retrieved 4 December 2016.
- ↑ Lai, Kang Yiu; Ng, Wing Yiu George; Cheng, Fan Fanny (28 November 2014). "Human Ebola virus infection in West Africa: a review of available therapeutic agents that target different steps of the life cycle of Ebola virus". Infectious Diseases of Poverty 3: 43. doi:10.1186/2049-9957-3-43. ISSN 2049-9957. PMID 25699183.
- ↑ 37.0 37.1 "Ebola – A Growing Threat?". N. Engl. J. Med. 371 (15): 1375–1378. May 2014. doi:10.1056/NEJMp1405314. PMID 24805988.
- ↑ Ng, S.; Cowling, B. (2014). "Association between temperature, humidity and ebolavirus disease outbreaks in Africa, 1976 to 2014". Eurosurveillance 19 (35): 20892. doi:10.2807/1560-7917.ES2014.19.35.20892. PMID 25210981.
- ↑ "Clinical care for survivors of Ebola virus disease". World Health Organization. 2016. http://apps.who.int/iris/bitstream/10665/204235/1/WHO_EVD_OHE_PED_16.1_eng.pdf?ua=1.
- ↑ "Clinical Aspects of Ebola Virus Disease at the Ngaliema Hospital, Kinshasa, Zaire, 1976". European Network for Diagnostics of "Imported" Viral Diseases (ENIVD). http://www.itg.be/internet/ebola/ebola-12.htm.
- ↑ Bardi, Jason Socrates. "Death Called a River". http://www.scripps.edu/newsandviews/e_20020114/ebola1.html.
- ↑ name: S. Reardan.; N Engl. J Med. (2014) " The first nine months of the epidemic and projection, Ebola virus disease in west Africa". archive of Ebola Response Team. 511(75.11):520
- ↑ Gina Kolata (Oct 30, 2014). "Genes Influence How Mice React to Ebola, Study Says in 'Significant Advance'". New York Times. https://www.nytimes.com/2014/10/31/health/genes-influence-ebola-infections-in-mice-study-suggests.html.
- ↑ Rasmussen, Angela L. (Oct 30, 2014). "Host genetic diversity enables Ebola hemorrhagic fever pathogenesis and resistance". Science 346 (6212): 987–991. doi:10.1126/science.1259595. PMID 25359852. Bibcode: 2014Sci...346..987R.
- ↑ Henao-Restrepo, Ana Maria (22 December 2016). "Efficacy and effectiveness of an rVSV-vectored vaccine in preventing Ebola virus disease: final results from the Guinea ring vaccination, open-label, cluster-randomised trial (Ebola Ça Suffit!)". The Lancet 389 (10068): 505–518. doi:10.1016/S0140-6736(16)32621-6. PMID 28017403.
- ↑ Berlinger, Joshua (22 December 2016). "Ebola vaccine gives 100% protection, study finds". CNN. http://www.cnn.com/2016/12/22/health/ebola-vaccine-study/index.html.
- ↑ "First FDA-approved vaccine for the prevention of Ebola virus disease, marking a critical milestone in public health preparedness and response". 19 December 2019. https://www.fda.gov/news-events/press-announcements/first-fda-approved-vaccine-prevention-ebola-virus-disease-marking-critical-milestone-public-health.
- ↑ Brown, Rob (2014-07-18). "The virus detective who discovered Ebola". BBC News. https://www.bbc.com/news/magazine-28262541.
- ↑ "Isolation and partial characterisation of a new virus causing haemorrhagic fever in Zambia". Lancet 309 (8011): 569–571. 1977. doi:10.1016/s0140-6736(77)92000-1. PMID 65661.
- ↑ "Family Filoviridae". Virus Taxonomy – Seventh Report of the International Committee on Taxonomy of Viruses. San Diego: Academic Press. 2000. pp. 539–548. ISBN 978-0123702005.
- ↑ Pringle, C. R. (1998). "Virus taxonomy – San Diego 1998". Archives of Virology 143 (7): 1449–1459. doi:10.1007/s007050050389. PMID 9742051.
- ↑ "Family Filoviridae". Virus Taxonomy – Eighth Report of the International Committee on Taxonomy of Viruses. San Diego: Elsevier/Academic Press. 2005. pp. 645–653. ISBN 978-0123702005.
- ↑ Mayo, M. A. (2002). "ICTV at the Paris ICV: results of the plenary session and the binomial ballot". Archives of Virology 147 (11): 2254–2260. doi:10.1007/s007050200052.
- ↑ "Replace the species name Lake Victoria marburgvirus with Marburg marburgvirus in the genus Marburgvirus". http://talk.ictvonline.org/files/ictv_official_taxonomy_updates_since_the_8th_report/m/vertebrate-official/4171.aspx.
- ↑ International Committee on Taxonomy of Viruses. "Virus Taxonomy: 2013 Release". http://ictvonline.org/virusTaxonomy.asp.
- ↑ "Role of Ebola Virus Secreted Glycoproteins and Virus-Like Particles in Activation of Human Macrophages". Journal of Virology 79 (4): 2413–2419. 2005. doi:10.1128/JVI.79.4.2413-2419.2005. PMID 15681442.
- ↑ "Retinazone inhibits certain blood-borne human viruses including Ebola virus Zaire". Antiviral Chemistry & Chemotherapy 23 (5): 197–215. 2014. doi:10.3851/IMP2568. PMID 23636868.
- ↑ 58.0 58.1 Carroll, S.A. (2012). "Molecular Evolution of Viruses of the Family Filoviridae Based on 97 Whole-Genome Sequences". Journal of Virology 87 (5): 2608–2616. doi:10.1128/JVI.03118-12. PMID 23255795.
- ↑ 59.0 59.1 Li, Y.H. (2013). "Evolutionary history of Ebola virus". Epidemiology and Infection 142 (6): 1138–1145. doi:10.1017/S0950268813002215. PMID 24040779.
- ↑ 60.0 60.1 Wittmann TJ, Biek R, Hassanin A, Rouquet P, Reed P, Yaba P, Pourrut X, Real LA, Gonzalez JP, Leroy EM. "Isolates of Zaire ebolavirus from wild apes reveal genetic lineage and recombinants". Proc Natl Acad Sci U S A. 2007 Oct 23;104(43):17123–17127. Epub 2007 Oct 17. "Erratum" in: Proc Natl Acad Sci U S A. 2007 Dec 4;104(49):19656. PMID 17942693
- ↑ 61.0 61.1 "Outbreaks Chronology: Ebola Virus Disease" (in en-us). Ebola Hemorrhagic Feve. CDC. 2017-08-02. https://www.cdc.gov/vhf/ebola/outbreaks/history/chronology.html.
- ↑ Kava, Alex (October 2008) (in en). Fiction Book Review: 'Exposed' by Alex Kava, Author . Mira (332p). PublishersWeekly.com. ISBN 978-0778325574. https://www.publishersweekly.com/978-0-7783-2557-4. Retrieved 2021-11-07.
- ↑ Close, William T. (1995). Ebola: A Documentary Novel of Its First Explosion. New York: Ivy Books. ISBN 978-0804114325. OCLC 32753758. https://archive.org/details/eboladocumentary00clos. At Google Books.
- ↑ Grove, Ryan (2006-06-02). More about the people than the virus. https://www.amazon.com/Ebola-documentary-novel-first-explosion/dp/B007HEJSE6. Retrieved 2014-09-17.
- ↑ Close, William T. (2002). Ebola: Through the Eyes of the People. Marbleton, Wyoming: Meadowlark Springs Productions. ISBN 978-0970337115. OCLC 49193962. https://books.google.com/books?id=BX0gAQAAIAAJ. At Google Books.
- ↑ Pink, Brenda (2008-06-24). "A fascinating perspective". https://www.amazon.com/Ebola-Through-William-T-Close/product-reviews/0970337116.
- ↑ Preston, Richard (1995). The Hot Zone. New York: Anchor. ISBN 0385479565. OCLC 32052009.
- ↑ Clancy, Tom (1996). Executive Orders. New York: Putnam. ISBN 978-0399142185. OCLC 34878804. https://archive.org/details/executiveorders00clan.
- ↑ Stone, Oliver (1996-09-02). "Who's That in the Oval Office?". https://www.nytimes.com/books/97/08/17/bsp/20141.html.
Further reading
- Pacheco, Daniela Alexandra de Meneses Rocha; Rodrigues, Acácio Agostinho Gonçalves; Silva, Carmen Maria Lisboa da (October 2016). "Ebola virus – from neglected threat to global emergency state". Revista da Associação Médica Brasileira 62 (5): 458–467. doi:10.1590/1806-9282.62.05.458. PMID 27656857.
External links
| Wikimedia Commons has media related to Ebolavirus. |
- Ebolavirus molecular biology
- Ebolavirus proteins (PDB-101)
- ICTV Files and Discussions – Discussion forum and file distribution for the International Committee on Taxonomy of Viruses
- Genomic data on Ebola virus isolates and other members of the family Filoviridae
- ViralZone: Ebola-like viruses – Virological repository from the Swiss Institute of Bioinformatics
- Virus Pathogen Resource: Ebola Portal – Genomic and other research data about Ebola and other human pathogenic viruses
- The Ebola Virus 3D model of the Ebola virus, prepared by Visual Science, Moscow.
- FILOVIR – scientific resources for research on filoviruses
- "'Zaire ebolavirus'". https://www.ncbi.nlm.nih.gov/Taxonomy/Browser/wwwtax.cgi?mode=Info&id=186538.
- "'Ebola virus sp.'". https://www.ncbi.nlm.nih.gov/Taxonomy/Browser/wwwtax.cgi?mode=Info&id=205488.
Wikidata ☰ Q8064876 entry
 |