Biology:Glutamate-rich protein 4
Glutamate-rich protein 4 is encoded by the gene ERICH4 and can be otherwise known as chromosome 19 open reading frame 69 (C19orf69).[1] ERICH4 is highly conserved in mammals and exhibits overexpression in tissues of the kidneys, terminal ileum, and duodenum.[1][2] The function of ERICH4 has yet to be well understood by the scientific community but is suggested to contribute to immune inflammatory responses.
Gene
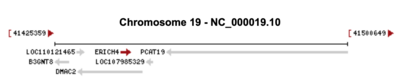
ERICH4 is located on the sense strand of 19q13.2 in humans, consists of 2,340 base pairs, and contains 2 exons.[3] ERICH4, on the sense strand, is located within DMAC2 and next PCAT19 and B3NT8 which are all on the antisense strand.[1]
Promoter & Predicted Transcription Factors (TF)
The promoter is predicted to begin 1,806 bp upstream from the 5' UTR and consists of 1,819 bp which overlaps with the coding sequence by 13 bp.[4]
| Matrix ID | TF Name | Genomic Position with Human ERICH4 Promoter | Strand of q19 | Matrix Similarity | Literature Supported Function |
|---|---|---|---|---|---|
| V$CEBPA.01 | CCAAT/Enhancer-binding Protein Alpha | 41,441,752-41,441,766 | Sense (+) | 0.962 | Recruit co-activators that in turn can open up chromatin structure or recruit basal transcription factors.[5][6] |
| O$VTATA.01 | Vertebrate TATA-binding Factor | 41,442,466-41,442,482 | Antisense (-) | 0.915 | Required for initiation of transcription and is associated with a variety of different transcription factors.[7] |
| V$SOX1.04 | SRY (Sex Determining Region)-Box 1 | 41,442,480-41,442,502 | Sense (+) | 0.801 | Involved in the regulation of embryonic development and in the determination of the cell fate.[8] |
| V$HSF1.04 | Heat Shock Factor 1 | 41,442,536-41,442,560 | Sense (+) | 0.769 | Activation in cellular stress.[9] |
| V$BTEB3.01 | Krueppel-like Factor 13 (KLF13) | 41,442,243-41,442,261 | Sense (+) | 0.934 | KLF13 knock-out mice show a defect in lymphocyte survival as KLF13 is a regulator of Bcl-xL expression.[10] |
| V$PAX5.01 | B-cell Specific Activator Protein | 41,442,955-41,442,983 | Sense (+) | 0.796 | Key role in B-lymphocyte development.[11] |
| V$GLI3.02 | GLI-Kruppel Family Member GLI3 | 41,443,115-41,443,131 | Sense (+) | 0.915 | Thought to play a role during embryogenesis.[12] |
| V$NR2F6.01 | Nuclear Receptor subfamily 2 group F member 6 (NR2F6) | 41,442,507-41,442,531 | Sense (+) | 0.851 | Transcriptional repressor of IL17 expression in Th17-differentiated CD4-positive T cells in-vitro and in-vivo.[13] |
| V$MAFB.01 | MAFB/Leucine Zipper Transcription Factor | 41,442,601-41,442,625 | Sense (+) | 0.923 | Regulation of lineage-specific hematopoiesis. Represses ETS1-mediated transcription of erythroid-specific genes in myeloid cells.[14] |
| V$AP4.03 | Activating Enhancer Binding Protein 4 (TFAP4) | 41,443,003-41,443,019 | Sense (+) | 0.993 | Regulates the expression of genes involved in the regulation of cellular proliferation, stemness, and epithelial-mesenchymal transition.[15] |
| V$EVI1.05 | Ecotropic Viral Integration Site 1 (EVI1) Encoded Factor | 41,442,583-41,442,599 | Sense (+) | 0.821 | Regulation of hematopoietic stem cell renewal. Controls several aspects of embryonic development.[16] |
| V$MRE.01 | Mineralcorticoid Receptor Response Element | 41,442,844-41,442,862 | Sense (+) | 0.939 | Involved in water electrolyte homeostasis, blood pressure regulation, inflammation, and fibrosis in the renocardiovascular system.[17] |
| V$ARE.03 | Androgene Receptor Binding Site, IR3 Sites | 41,442,844-41,442,862 | Antisense (-) | 0.946 | Ligand-dependent transcription factor that controls the expression of specific genes. The binding of the AR to its native ligands 5α-dihydrotestosterone (DHT) and testosterone initiates male sexual development and differentiation.[18] |
| V$ZNF217.01 | Zinc Finger Protein 217 | 41,443,023-41,443,035 | Sense (+) | 0.911 | Promotes cell proliferation and antagonizes cell death.[19] |
| V$RORA.02 | RAR-related Orphan Receptor Alpha, Homodimer DR5 Binding Site | 41,442,783-41,442,807 | Sense (+) | 0.831 | Possible role in lymphocyte development. Possible function in negatively regulating inflammation due to a report of positive relation in the expression of IKBa, a negative regulator of the NF-kB signaling pathway.[20] |
| V$STAT6.01 | Signal Transducer and Activator of Transcription 6 (STAT6) | 41,443,042-41,443,060 | Sense (+) | 0.961 | Plays a central role in exerting IL4-mediated responses.[21] |
| V$ZF5.01 | Zinc Finger/POZ Transcription Factor | 41,442,874-41,442,888 | Sense (+) | 0.957 | Role in development, oncogenesis, apoptosis, and transcription repression.[22] |
mRNA
The ERICH4 mRNA sequence is 955 nucleotides in length with a fold energy predicted as -139.80 kcal/mol with -0.258 energy/base.[23]
Alternative Splicing
ERICH4 has one different protein-encoding transcript variant, or isoform.[1]
| Name | mRNA Length (bp) | Protein Length (aa) | Mass (Da) |
|---|---|---|---|
| Glutamate-rich protein 4 | 955 | 130 | 14,447 |
| Glutamate-rich protein 4 isoform X1 | 1741 | 155 | N/A |
Protein
General Properties
The primary encoded protein consists of 130 amino acids and has a predicted molecular mass of 14.5 kDa and isoelectric point of 4 pI.[24] As suggested by the protein's name, glutamate-rich protein 4, the protein is most highly composed of glutamic acid amino acids at 17.7% of the protein's composition followed by leucine at 14.6%, and then proline at 9.2%.[25] ERICH4 has no positive or negative charge clusters.[25] The human protein has one identifiable mixed cluster from amino acid 91 to 116 with 3 positively-charged, 15 negatively-charged, and 8 neutral amino acids.[25] The same mixed cluster region in humans is frequently negative within ERICH4's orthologous proteins.[25] This protein contains no significant hydrophobic or transmembrane segments which are supported with comparison to five of ERICH4's orthologs (Graymouse lemur, Sheep, House mouse, African elephant, and Opossum).[25]
Domains
ERICH4 has one identified domain of unknown function, DUF4530, which is found in eukaryotes.[26] Proteins in this family are typically 140 amino acids in length and ERICH4 is a known human member of this family.[27]
Secondary Structure
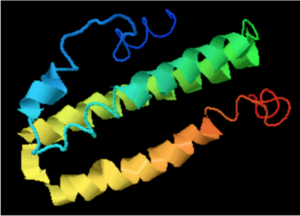
A cross-program analysis determines ERICH4 protein to be composed of five separated alpha helixes and five interspersing coils. The alpha helix segments span from amino acids 2-9, 21-24, 47-58, 61-94, and 104-111 in the protein sequence. ERICH4 is not predicted to contain beta-sheets.[28][29][30][31]
Tertiary Structure
Program analysis in SWISS-Model proposes a tertiary structure for ERICH4 by matching the protein against the template of NLRP6 with a sequence identity of 25.79%, sequence similarity of 0.30, and coverage of 0.43 for amino acids 43-92 in ERICH4.[32]
Post-translational Regulation
ERICH4 has proposed phosphorylation at serine amino acids 28 and 96 and amino acid 36, a threonine, by casein kinase II and protein kinase c, respectively.[33][34] ERICH4 is not predicted to be undergo a methionine cleavage or acetylation.[35]
Localization
This protein is predicted to be intracellular without any transmembrane regions. Sub-cellular localization is predicted to be mostly localized to the cytoplasm with a reliability score of 70.6 via the Reinhardt's method.[36] No significant O-GlcNAc site and N-myristoylation predictions.[37]
Tissue Expression
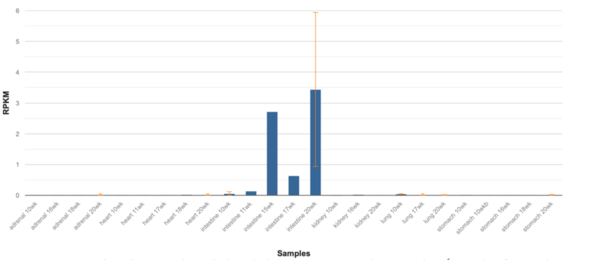
ERICH4’s highest levels of expression are within human tissue of the duodenum and small intestine, followed by the kidneys.[1][38] Notably, expression within the small intestines is highest in the twentieth week of human fetal development.[1][39] Within a representative set of mouse (Mus musculus) tissues, Erich4 is most highly expressed within the kidneys, followed by and in decreasing expression, the large intestines, adult duodenum, and adult small intestine.[40] The Sigma-Aldrich antibody product, HPA042632, derived from rabbit, has a strong granular cytoplasmic positivity in cytoplasmic structure in glandular cells (goblet cells) of the rectum.[41]
Abnormal Tissue Expression
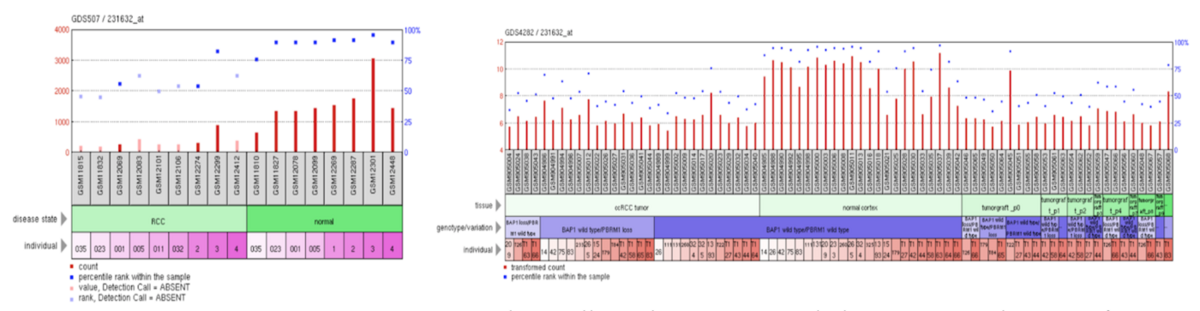
ERICH4 has high expression within normal tissue and low-to-medium expression with renal cell carcinoma tissue.[42]
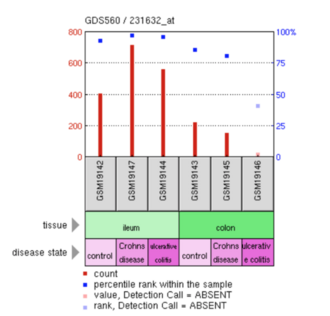
An analysis examining ERICH4 was reviewed in tissues of the ileum and colon that were either normal or afflicted with Crohn's disease or ulcerative colitis. ERICH4 had high (~90%) expression within the ileum for all states (normal/control, Crohn's disease, and ulcerative colitis).[43] ERICH4 also has a higher expression in Crohn's disease than in either normal tissue or ulcerative colitis.
Function
The function of ERICH4 has yet to be well understood by the scientific community and therefore, requires further research.
Interactions
According to STRING analysis, ERICH4 has multiple predicted interactions with other proteins including proteins with associated immune function and expression within the gastrointestinal tract or testes from textmining.[44] No experimentally confirmed protein interactions yet.
| Predicted Partner Protein | Score | Associated Functions |
|---|---|---|
| Tetratricopeptide Repeat Domain 29 (TTC29) | 0.680 | Shown to be significantly upregulated during wound healing of human masticatory mucosa.[45] |
| Transmembrane Protein 184A (TMEM184A) | 0.552 | Functions as a heparin receptor and mediates anti-inflammatory responses of ECs involving decreased JNK and p38 activity.[46] |
| Insulin-like Growth Factor binding protein Acid Labile Subunit (IGFALS) | 0.509 | Serum protein that binds insulin-like growth factors, increasing their half-life and their vascular localization.[47] |
| Serine Peptidase Inhibitor, Kazal type 4 (SPINK4) | 0.500 | Has been shown to exhibit Celiac disease pathology-related differential gene expression, likely derived from altered goblet cell activity.[48] |
| Protein Disulfide-Isomerase-Like protein of the Testis (PDILT) | 0.497 | Catalyzes protein folding and thiol-disulfide interchange reactions. This protein lacks oxidoreductase activity in vitro and is suspected to function as a chaperone.[49] |
Homology
Paralogs
No human paralogs were found for the gene.[2]
Orthologs
Orthologs have been identified in most mammals for which complete genome data is available. Notably, ERICH4 orthologs are only present in placental and marsupial mammals but absent in monotremes.[2] The most distant ortholog was identified in the gray short-tailed opossum which is a marsupial mammal.[2]
No significant similarities were found in the vertebrates Aves, Reptilia, Amphibia, Chondrichthyes, Osteichthyes or Agnatha. Searching to exclude vertebrates in BLAST and BLAT produced no significant ortholog findings for invertebrates, fungi, and bacteria.[2]
| Species | Common Name | NCBI Accession Number | Sequence Length (AA) | Millions of Years since LCA | % Identity | % Similarity | Taxonomic Group |
|---|---|---|---|---|---|---|---|
| Homo sapiens | Human | NP_001123986.1 | 130 | --- | 100 | 100 | Primates |
| Microcebus murinus | Gray mouse lemur | XP_012616209.1 | 179 | 73 | 72 | 80 | Primates |
| Tupaia chinensis | Northern treeshrew | XP_006165343.1 | 137 | 85 | 63 | 72 | Scandentia |
| Mus pahari | Gairdner's shrewmouse | XP_021075502.1 | 141 | 88 | 57 | 63 | Rodentia |
| Meriones unguiculatus | Mongolian gerbil | XP_021519873.1 | 141 | 88 | 60 | 64 | Rodentia |
| Rattus norvegicus | Brown rat | NP_001102923.1 | 147 | 88 | 61 | 65 | Rodentia |
| Mus musculus | House mouse | NP_001034332.2 | 140 | 88 | 62 | 71 | Rodentia |
| Microtus ochrogaster | Prairie vole | XP_005361243.1 | 140 | 88 | 62 | 71 | Rodentia |
| Erinaceus europaeus | European hedgehog | XP_007536664.1 | 129 | 94 | 61 | 68 | Eulipotyphla |
| Orcinus orca | Killer whale | XP_004271419.2 | 121 | 94 | 62 | 72 | Cetacea |
| Physeter catodon | Sperm whale | XP_007128192.1 | 121 | 94 | 64 | 72 | Cetacea |
| Desmodus rotundus | Common vampire bat | XP_024433457.1 | 180 | 94 | 66 | 73 | Chiroptera |
| Ovis aries | Sheep | XP_012045823.2 | 131 | 94 | 69 | 75 | Artiodactyla |
| Bos taurus | Cattle | XP_002695042.1 | 131 | 94 | 69 | 75 | Artiodactyla |
| Pteropus alecto | Black flying fox | XP_006910763.1 | 133 | 94 | 71 | 78 | Chiroptera |
| Hipposideros armiger | Great roundleaf bat | XP_019488166.1 | 134 | 94 | 71 | 79 | Chiroptera |
| Loxodonta africana | African bush elephant | XP_003420798.1 | 127 | 102 | 58 | 66 | Proboscidea |
| Monodelphis domestica | Gray short-tailed opossum | XP_007492011.1 | 106 | 160 | 45 | 61 | Didelphimorphia |
| Phascolarctos cinereus | Koala | XP_020834126.1 | 109 | 160 | 48 | 63 | Diprotodontia |
| Vombatus ursinus | Common wombat | XP_027701859.1 | 109 | 160 | 49 | 64 | Diprotodontia |
Molecular Evolution
The m value, or number of corrected amino acid changes per 100 residues, for the gene ERICH4 was plotted against the divergence of species in millions of years. When compared to the data of hemoglobin, fibrinogen alpha chain, and cytochrome C, it was determined that the gene has the closest progression to fibrinogen alpha chain, suggesting a relatively rapid pace of evolution. M values for ERICH4 were derived from percentage of identity of species protein sequences compared to the human sequence using the formula derived from the Molecular Clock Hypothesis.
References
- ↑ 1.0 1.1 1.2 1.3 1.4 1.5 "ERICH4 glutamate rich 4 [Homo sapiens (human)"]. https://www.ncbi.nlm.nih.gov/gene/100170765.
- ↑ 2.0 2.1 2.2 2.3 2.4 "ERICH4 Orthologs". https://blast.ncbi.nlm.nih.gov/Blast.cgi.
- ↑ "ERICH4 Gene (Protein Coding)". https://www.genecards.org/cgi-bin/carddisp.pl?gene=ERICH4&keywords=erich4. Retrieved 7 February 2019.
- ↑ "Promoter of ERICH4". https://www.genomatix.de/cgi-bin/eldorado/eldorado.pl?s=a3287ff4267bce3176faefe29351c093;RESULT=ERICH4.
- ↑ Kovaks K, Steinmann M, Magistretti P, Halfon O, Cardinaux J (2003). "CCAAT/Enhancer-binding Protein Family Members Recruit the Coactivator CREB-binding Protein and Trigger Its Phosphorylation". The Journal of Biological Chemistry 278 (38): 36959–36965. doi:10.1074/jbc.M303147200. PMID 12857754.
- ↑ Ramji D.P., Foka P. (2002). "CCAAT/enhancer-binding proteins: structure, function and regulation". Biochemical Journal 365 (3): 561–575. doi:10.1042/BJ20020508. PMID 12006103.
- ↑ McDowall, Jennifer. "TATA-binding proteins". https://www.ebi.ac.uk/interpro/potm/2005_7/Page1.htm. Retrieved 6 May 2019.
- ↑ "SRY Gene". https://ghr.nlm.nih.gov/gene/SRY.
- ↑ Vihervaara A, Sistonen L (2014). "HSF1 at a glance". Journal of Cell Science 127 (2): 261–266. doi:10.1242/jcs.132605. PMID 24421309.
- ↑ Zhou M, McPherson L, Feng D, Song A, Dong C, Lyu SC, Zhou L, Shi X, Ahn YT, Wang D, Clayberger C, Krensky AM (May 2007). "Kruppel-like transcription factor 13 regulates T lymphocyte survival in vivo.". Journal of Immunology 178 (9): 5496–504. doi:10.4049/jimmunol.178.9.5496. PMID 17442931.
- ↑ Laszlo Krenacs, Andreas W. Himmelmann, Leticia Quintanilla-Martinez, Thierry Fest, Agostino Riva, Axel Wellmann, Eniko Bagdi, John H. Kehrl, Elaine S. Jaffe, and Mark Raffeld (August 15, 1998). "Transcription Factor B-Cell–Specific Activator Protein (BSAP) Is Differentially Expressed in B Cells and in Subsets of B-Cell Lymphomas". Blood 92 (4): 1308–16. doi:10.1182/blood.V92.4.1308. PMID 9694719. http://www.bloodjournal.org/cgi/pmidlookup?view=long&pmid=9694719.
- ↑ "GLI family zinc finger 3 (GLI3) [Homo sapiens"]. https://www.ncbi.nlm.nih.gov/gene/2737.
- ↑ Hermann-Kleiter N, Gruber T, Lutz-Nicoladoni C, Thuille N, Fresser F, Labi V, Schiefermeier N, Warnecke M, Huber L, Villunger A, Eichele G, Kaminski S, Baier G (2008). "The nuclear orphan receptor NR2F6 suppresses lymphocyte activation and T helper 17-dependent autoimmunity.". Immunity 29 (2): 205–216. doi:10.1016/j.immuni.2008.06.008. PMID 18701084.
- ↑ "MAF bZIP transcription factor B (MAFB) [Homo sapiens"]. https://www.ncbi.nlm.nih.gov/gene/9935.
- ↑ Lupia M, Angiolini F, Bertalot G, Freddi S, Sachsenmeier KF, Chisci E, Kutryb-Zajac B, Confalonieri S, Smolenski RT, Giovannoni R, Colombo N, Bianchi F, Cavallaro U. (2018). "CD73 Regulates Stemness and Epithelial-Mesenchymal Transition in Ovarian Cancer-Initiating Cells.". Stem Cell Reports 10 (4): 1412–1425. doi:10.1016/j.stemcr.2018.02.009. PMID 29551673.
- ↑ Yuasa H, Oike Y, Iwama A, Nishikata I, Sugiyama D, Perkins A, Mucenski ML, Suda T, Morishita K. (June 1, 2005). "Oncogenic transcription factor Evi1 regulates hematopoietic stem cell proliferation through GATA-2 expression.". EMBO Journal 24 (11): 1976–87. doi:10.1038/sj.emboj.7600679. PMID 15889140.
- ↑ Meinel S, Ruhs S, Schumann K, Strätz N, Trenkmann K, Schreier B, Grosse I, Keilwagen J, Gekle M, Grossmann C. (September 2013). "Mineralocorticoid receptor interaction with SP1 generates a new response element for pathophysiologically relevant gene expression.". Nucleic Acids Research 41 (17): 8045–60. doi:10.1093/nar/gkt581. PMID 23821666.
- ↑ Tan MH, Li J, Xu E, Melcher K, Yong E (2015). "Androgen receptor: structure, role in prostate cancer and drug discovery". Acta Pharmacologica Sinica 36 (1): 3–23. doi:10.1038/aps.2014.18. PMID 24909511.
- ↑ Guiqing Huang, Sheryl Krig, David Kowbel, Hongmei Xu, Bill Hyun, Stas Volik, Burt Feuerstein, Gordon B. Mills, David Stokoe, Paul Yaswen, Colin Collins (November 2005). "ZNF217 suppresses cell death associated with chemotherapy and telomere dysfunction". Human Molecular Genetics 14 (21): 3219–3225. doi:10.1093/hmg/ddi352. PMID 16203743.
- ↑ Delerive P, Monté D, Dubois G, Trottein F, Fruchart-Najib J, Mariani J, Fruchart JC, Staels B. (January 15, 2001). "The orphan nuclear receptor ROR alpha is a negative regulator of the inflammatory response.". EMBO Reports 2 (1): 42–48. doi:10.1093/embo-reports/kve007. PMID 11252722.
- ↑ "signal transducer and activator of transcription 6 (STAT6) [Homo sapiens"]. https://www.ncbi.nlm.nih.gov/gene/6778.
- ↑ Lee DK, Suh D, Edenberg HJ, Hur MW. (2002). "POZ domain transcription factor, FBI-1, represses transcription of ADH5/FDH by interacting with the zinc finger and interfering with DNA binding activity of Sp1". Journal of Biological Chemistry 277 (30): 2671–8. doi:10.1074/jbc.M202078200. PMID 12004059.
- ↑ "ERICH4". https://genome.ucsc.edu/cgi-bin/hgGene?hgg_gene=ENST00000378187.2&hgg_prot=ENST00000378187.2&hgg_chrom=chr19&hgg_start=41443157&hgg_end=41444765&hgg_type=knownGene&db=hg38&hgsid=710121421_MGTr8YVUFdKNo5F1OFLRzeUmXRYf. Retrieved 1 March 2019.
- ↑ "Compute pI/Mw tool, ERICH4". https://web.expasy.org/compute_pi/.
- ↑ 25.0 25.1 25.2 25.3 25.4 "ERICH4 Sequence Composition". https://web.expasy.org/compute_pi/.
- ↑ "Domains/Motifs of ERICH4". https://www.genome.jp/tools/motif/.
- ↑ "DUF4530". https://www.ncbi.nlm.nih.gov/Structure/cdd/cl20913.
- ↑ "ERICH4 Secondary Structure". https://npsa-prabi.ibcp.fr/cgi-bin/secpred_gor4.pl.
- ↑ Ethan Wolf, Peter S. Kim, and Bonnie Berger (June 1997). "MultiCoil: A Program for Predicting Two- and Three-Stranded Coiled Coils". Protein Science 6 (6): 1179–1189. doi:10.1002/pro.5560060606. PMID 9194178.
- ↑ "Proposed Tertiary Structure for ERICH4". http://www.sbg.bio.ic.ac.uk/phyre2/phyre2_output/ea0aa5d9472abd66/summary.html.
- ↑ "Proposed Tertiary Structure for ERICH4". https://zhanglab.ccmb.med.umich.edu/I-TASSER/output/S461040/.
- ↑ "SWISS-Model of ERICH4". https://swissmodel.expasy.org/interactive/Z39nep/models/.
- ↑ "Phosphorylation Sites for ERICH4". http://www.cbs.dtu.dk/cgi-bin/webface2.fcgi?jobid=5CB8DE2600006CE714A1C619&wait=20.
- ↑ "SIB Motif Scan". https://myhits.isb-sib.ch/cgi-bin/motif_scan.
- ↑ "SIB Terminus, N-Terminal PTM Prediction". http://terminus.unige.ch/predict.
- ↑ "ERICH4 Sub-cellular Localization". https://psort.hgc.jp/cgi-bin/runpsort.pl.[yes|permanent dead link|dead link}}]
- ↑ "YinOYang GlcNAc Predictor". http://www.cbs.dtu.dk/cgi-bin/webface2.fcgi?jobid=5CB9274A000030675E44B982&wait=20.
- ↑ Fagerberg L, Hallström BM, Oksvold P, Kampf C, Djureinovic D, Odeberg J, Habuka M, Tahmasebpoor S, Danielsson A, Edlund K, Asplund A, Sjöstedt E, Lundberg E, Szigyarto CA, Skogs M, Takanen JO, Berling H, Tegel H, Mulder J, Nilsson P, Schwenk JM, Lindskog C, Danielsson F, Mardinoglu A, Sivertsson A, von Feilitzen K, Forsberg M, Zwahlen M, Olsson I, Navani S, Huss M, Nielsen J, Ponten F, Uhlén M. (2013). "Analysis of the human tissue-specific expression by genome-wide integration of transcriptomics and antibody-based proteomics.". Molecular & Cellular Proteomics 13 (2): 397–406. doi:10.1074/mcp.M113.035600. PMID 24309898.
- ↑ Szabo L, Morey R, Palpant NJ, Wang PL, Afari N, Jiang C, Parast MM, Murry CE, Laurent LC, Salzman J. (June 16, 2016). Statistically based splicing detection reveals neural enrichment and tissue-specific induction of circular RNA during human fetal development..
- ↑ Yue F, Cheng Y, Breschi A, Vierstra J, Wu W, Ryba T, Sandstrom R, Ma Z, Davis C, Pope BD, Shen Y, Pervouchine DD, Djebali S, Thurman RE, Kaul R, Rynes E, Kirilusha A, Marinov GK, Williams BA, Trout D, Amrhein H, Fisher-Aylor K, Antoshechkin I, DeSalvo G, See LH, Fastuca M, Drenkow J, Zaleski C, Dobin A, Prieto P, Lagarde J, Bussotti G, Tanzer A, Denas O, Li K, Bender MA, Zhang M, Byron R, Groudine MT, McCleary D, Pham L, Ye Z, Kuan S, Edsall L, Wu YC, Rasmussen MD, Bansal MS, Kellis M, Keller CA, Morrissey CS, Mishra T, Jain D, Dogan N, Harris RS, Cayting P, Kawli T, Boyle AP, Euskirchen G, Kundaje A, Lin S, Lin Y, Jansen C, Malladi VS, Cline MS, Erickson DT, Kirkup VM, Learned K, Sloan CA, Rosenbloom KR, Lacerda de Sousa B, Beal K, Pignatelli M, Flicek P, Lian J, Kahveci T, Lee D, Kent WJ, Ramalho Santos M, Herrero J, Notredame C, Johnson A, Vong S, Lee K, Bates D, Neri F, Diegel M, Canfield T, Sabo PJ, Wilken MS, Reh TA, Giste E, Shafer A, Kutyavin T, Haugen E, Dunn D, Reynolds AP, Neph S, Humbert R, Hansen RS, De Bruijn M, Selleri L, Rudensky A, Josefowicz S, Samstein R, Eichler EE, Orkin SH, Levasseur D, Papayannopoulou T, Chang KH, Skoultchi A, Gosh S, Disteche C, Treuting P, Wang Y, Weiss MJ, Blobel GA, Cao X, Zhong S, Wang T, Good PJ, Lowdon RF, Adams LB, Zhou XQ, Pazin MJ, Feingold EA, Wold B, Taylor J, Mortazavi A, Weissman SM, Stamatoyannopoulos JA, Snyder MP, Guigo R, Gingeras TR, Gilbert DM, Hardison RC, Beer MA, Ren B. (2014). "A comparative encyclopedia of DNA elements in the mouse genome.". Nature 515 (7527): 355–364. doi:10.1038/nature13992. PMID 25409824. Bibcode: 2014Natur.515..355..
- ↑ "The Human Protein Atlas, Antibody information". https://www.proteinatlas.org/ENSG00000204978-ERICH4/antibody.
- ↑ Lenburg ME, Liou LS, Gerry NP, Frampton GM, Cohen HT, Christman MF. (2003). "Previously unidentified changes in renal cell carcinoma gene expression identified by parametric analysis of microarray data.". BMC Cancer 3: 31. doi:10.1186/1471-2407-3-31. PMID 14641932.
- ↑ Moehle C, Ackermann N, Langmann T, Aslanidis C, Kel A, A, Kel-Margoulis O, Schmitz-Madry A, Zahn A, Stremmel W, Schmitz G. (2006). "Aberrant intestinal expression and allelic variants of mucin genes associated with inflammatory bowel disease". J. Mol. Med. 84 (12): 1055–66. doi:10.1007/s00109-006-0100-2. PMID 17058067.
- ↑ "Protein Interactions for ERICH4". https://string-db.org.
- ↑ Wang, Y, Tatakis, DN. (2017). "Human gingiva transcriptome during wound healing". J Clin Periodontol 44 (4): 394–402. doi:10.1111/jcpe.12669. PMID 28005267.
- ↑ Farwell SL, Kanyi D, Hamel M, Slee JB, Miller EA, Cipolle MD, Lowe-Krentz LJ. (2016). "Heparin Decreases in Tumor Necrosis Factor α (TNFα)-induced Endothelial Stress Responses Require Transmembrane Protein 184A and Induction of Dual Specificity Phosphatase 1.". Journal of Biological Chemistry 291 (10): 5342–5354. doi:10.1074/jbc.M115.681288. PMID 26769965.
- ↑ "IGFALS insulin like growth factor binding protein acid labile subunit [Homo sapiens (human)"]. https://www.ncbi.nlm.nih.gov/gene/3483. Retrieved 6 May 2019.
- ↑ Wapenaar MC, Monsuur AJ, Poell J, van 't Slot R, Meijer JW, Meijer GA, Mulder CJ, Mearin ML, Wijmenga C. (2007). "The SPINK gene family and celiac disease susceptibility.". Immunogenetics 59 (5): 349–357. doi:10.1007/s00251-007-0199-5. PMID 17333166.
- ↑ "PDILT protein disulfide isomerase like, testis expressed [Homo sapiens (human)"]. https://www.ncbi.nlm.nih.gov/gene/204474.
 |