Biology:Alloherpesviridae
| Alloherpesviridae | |
|---|---|
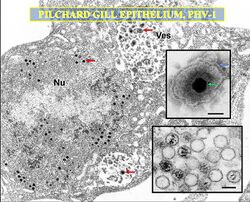
| |
| TEM micrograph on an alloherpesvirus, 'Pilchard herpesvirus' | |
| Virus classification | |
| (unranked): | Virus |
| Realm: | Duplodnaviria |
| Kingdom: | Heunggongvirae |
| Phylum: | Peploviricota |
| Class: | Herviviricetes |
| Order: | Herpesvirales |
| Family: | Alloherpesviridae |
| Genera | |
|
See text | |
Alloherpesviridae is a family of viruses in the order Herpesvirales. This family includes the species that infect fish and amphibians. Phylogenetic studies have confirmed the validity of this family and suggest that it may be divided into two clades: one consisting of viruses from cyprinid and anguillid hosts and the other of viruses from ictalurid, salmonid, acipenserid, and ranid hosts.[1] There are currently 13 species in this family, divided among four genera. A disease associated with this family includes channel catfish disease.[2][3]
Taxonomy
Alloherpesviridae was established as a family in 2005.
Genera
The family contains the following four genera:[3]
Structure
Viruses in Alloherpesviridae are enveloped, with icosahedral and spherical to pleomorphic geometries, and T=16 symmetry. The diameter is around 150–200 nm. Genomes are linear and non-segmented, around 134-248kb in length. [2]
| Genus | Structure | Symmetry | Capsid | Genomic arrangement | Genomic segmentation |
|---|---|---|---|---|---|
| Ictalurivirus | Spherical pleomorphic | T=16 | Enveloped | Circular | Monopartite |
| Cyprinivirus | Spherical pleomorphic | T=16 | Enveloped | Linear | Monopartite |
| Batrachovirus | Spherical pleomorphic | T=16 | Enveloped | Linear | Monopartite |
| Salmonivirus | Spherical pleomorphic | T=16 | Enveloped | Linear | Monopartite |
Life cycle
Viral replication is nuclear, and is lysogenic. Entry into the host cell is achieved by attachment of the viral glycoproteins to host receptors, which mediates endocytosis. DNA-templated transcription is the method of transcription. Fish serve as the natural host. Transmission routes are passive diffusion.[2]
| Genus | Host details | Tissue tropism | Entry details | Release details | Replication site | Assembly site | Transmission |
|---|---|---|---|---|---|---|---|
| Ictalurivirus | Fish | None | Glycoprotiens | Budding | Nucleus | Nucleus | Passive diffusion |
| Cyprinivirus | Fresh water eel | None | Glycoprotiens | Budding | Cytoplasm | Cytoplasm | Passive diffusion |
| Batrachovirus | Frogs | None | Glycoprotiens | Budding | Cytoplasm | Cytoplasm | Passive diffusion |
| Salmonivirus | Salmonidae | None | Glycoprotiens | Budding | Nucleus | Nucleus | Passive diffusion |
Genomics
Several genomes have been sequenced.[4] Cyprinid herpesviruses 1, 2 and 3 (CyHV1, CyHV2 and CyHV3) cause disease in common carp, goldfish and koi. Their genomes are 291144, 290304 and 295146 base pairs in size. The overall organisation common to all three and consists of a unique central region flanked by a direct repeat at each end. 137, 150, and 155 unique, functional protein coding genes are present in the unique regions: of these six, four, and eight are duplicated in the terminal repeat. The genomes share 120 orthologous genes in a largely colinear arrangement. Up to 55 of these latter genes are also conserved in the other member of Cyprinivirus; Anguillid herpesvirus 1. CyHV1, CyHV2, and CyHV3 have five, six and five families of paralogous genes.
References
- ↑ "Phylogenetic relationships in the family Alloherpesviridae". Dis. Aquat. Org. 84 (3): 179–94. 2009. doi:10.3354/dao02023. PMID 19565695.
- ↑ 2.0 2.1 2.2 "Viral Zone". ExPASy. http://viralzone.expasy.org/all_by_species/528.html.
- ↑ 3.0 3.1 "Virus Taxonomy: 2019 Release". International Committee on Taxonomy of Viruses. https://ictv.global/taxonomy.
- ↑ "Comparative genomics of carp herpesviruses". J. Virol. 87 (5): 2908–22. 2013. doi:10.1128/JVI.03206-12. PMID 23269803.
Further reading
- Inoue, Saga, Aikawa, Kumagai, Shimada, Kawaguchi, Naruse, Morishita, Koga, Takeda (2017). "Complete fusion of a transposon and herpesvirus created the Teratorn mobile element in medaka fish". Nature Communications 8 (1): 551. doi:10.1038/s41467-017-00527-2. PMID 28916771. Bibcode: 2017NatCo...8..551I.
External links
Wikidata ☰ Q3612626 entry
 |

