Biology:Coronaviridae
| Coronaviridae | |
|---|---|
| |
| |
| Diagram, electron micrograph, and genome of coronavirus types. | |
| Virus classification | |
| (unranked): | Virus |
| Realm: | Riboviria |
| Kingdom: | Orthornavirae |
| Phylum: | Pisuviricota |
| Class: | Pisoniviricetes |
| Order: | Nidovirales |
| Suborder: | Cornidovirineae |
| Family: | Coronaviridae |
| Subfamilies and genera | |
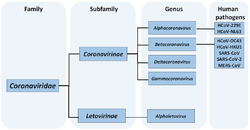
Coronaviridae is a family of enveloped, positive-strand RNA viruses which infect amphibians, birds, and mammals. The group includes the subfamilies Letovirinae and Orthocoronavirinae; the members of the latter are known as coronaviruses.
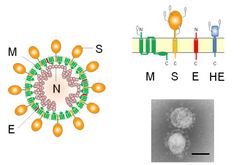
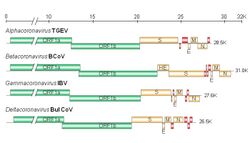
The viral genome is 26–32 kilobases in length. The particles are typically decorated with large (~20 nm), club- or petal-shaped surface projections (the "peplomers" or "spikes"), which in electron micrographs of spherical particles create an image reminiscent of the solar corona.[1][2][3]
Virology
The 5' and 3' ends of the genome have a cap and poly(A) tract, respectively. The viral envelope, obtained by budding through membranes of the endoplasmic reticulum (ER) or Golgi apparatus, invariably contains two virus-specified glycoprotein species, known as the spike (S) and membrane (M) proteins. The spike protein makes up the large surface projections (sometimes known as peplomers), while the membrane protein is a triple-spanning transmembrane protein. Toroviruses and a select subset of coronaviruses (in particular the members of subgroup A in the genus Betacoronavirus) possess, in addition to the peplomers composed of S, a second type of surface projections composed of the hemagglutinin-esterase protein. Another important structural protein is the phosphoprotein nucleocapsid protein (N), which is responsible for the helical symmetry of the nucleocapsid that encloses the genomic RNA.[4] The fourth and smallest viral structural protein is known as the envelope protein (E), thought to be involved in viral budding.[5]
Genetic recombination can occur when at least two viral genomes are present in the same infected host cell. RNA recombination appears to be a major driving force in coronavirus evolution. Recombination can determine genetic variability within a CoV species, the capability of a CoV species to jump from one host to another and, infrequently, the emergence of a novel CoV.[6] The exact mechanism of recombination in CoVs is not known, but likely involves template switching during genome replication.[6]
Taxonomy
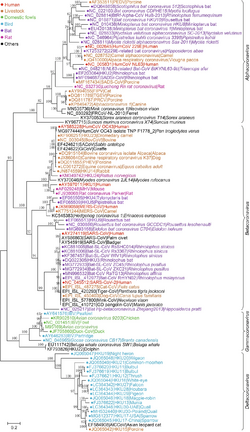
The family Coronaviridae is organized in 2 sub-families, 5 genera, 26 sub-genera, and 46 species.[7] Additional species are pending or tentative.[8]
- Coronaviridae
- Orthocoronavirinae[9]
- Letovirinae
- Alphaletovirus
- Milecovirus
- Microhyla letovirus 1
- Milecovirus
- Alphaletovirus
Coronavirus is the common name for Coronaviridae and Orthocoronavirinae, also called Coronavirinae.[10][11] Coronaviruses cause diseases in mammals and birds. In humans, the viruses cause respiratory infections. Four human coronaviruses cause typically minor symptoms of a common cold, while three are known to cause more serious illness and can be lethal: SARS-CoV-1, which causes SARS; MERS-CoV, which causes MERS; and SARS-CoV-2, which causes COVID-19.[12] Symptoms vary in other species: in chickens, they cause an upper respiratory disease, while in cows and pigs coronaviruses cause diarrhea. Other than for SARS-CoV-2, there are no vaccines or antiviral drugs to prevent or treat human coronavirus infections. They are enveloped viruses with a positive-sense single-stranded RNA genome and a nucleocapsid of helical symmetry. The genome size of coronaviruses ranges from approximately 26 to 32 kilobases, among the largest for an RNA virus (second only to a 41-kb nidovirus recently discovered in planaria).[13]
- Orthocoronavirinae
- Alphacoronavirus
- Colacovirus
- Bat coronavirus CDPHE15
- Decacovirus
- Bat coronavirus HKU10
- Rhinolophus ferrumequinum alphacoronavirus HuB-2013
- Duvinacovirus
- Human coronavirus 229E
- Luchacovirus
- Lucheng Rn rat coronavirus
- Minacovirus
- Mink coronavirus 1
- Minunacovirus
- Miniopterus bat coronavirus 1
- Miniopterus bat coronavirus HKU8
- Myotacovirus
- Myotis ricketti alphacoronavirus Sax-2011
- Nyctacovirus
- Nyctalus velutinus alphacoronavirus SC-2013
- Pipistrellus kuhlii coronavirus 3398
- Pedacovirus
- Porcine epidemic diarrhea virus
- Scotophilus bat coronavirus 512
- Rhinacovirus
- Rhinolophus bat coronavirus HKU2
- Setracovirus
- Human coronavirus NL63
- NL63-related bat coronavirus strain BtKYNL63-9b
- Soracovirus
- Sorex araneus coronavirus T14
- Sunacovirus
- Suncus murinus coronavirus X74
- Tegacovirus
- Colacovirus
- Betacoronavirus
- Embecovirus
- Hibecovirus
- Bat Hp-betacoronavirus Zhejiang2013
- Merbecovirus
- Nobecovirus
- Sarbecovirus
- Severe acute respiratory syndrome–related coronavirus
- Severe acute respiratory syndrome coronavirus (SARS-CoV)
- Severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2, cause of COVID-19)
- Severe acute respiratory syndrome–related coronavirus
- Gammacoronavirus
- Brangacovirus
- Goose coronavirus CB17
- Cegacovirus
- Beluga whale coronavirus SW1
- Igacovirus
- Avian coronavirus
- Avian coronavirus 9203
- Duck coronavirus 2714
- Brangacovirus
- Deltacoronavirus
- Andecovirus
- Wigeon coronavirus HKU20
- Buldecovirus
- Bulbul coronavirus HKU11
- Common moorhen coronavirus HKU21
- Coronavirus HKU15
- Munia coronavirus HKU13
- White-eye coronavirus HKU16
- Herdecovirus
- Night heron coronavirus HKU19
- Andecovirus
- Alphacoronavirus
References
- ↑ Alfarouk, Khalid O.; AlHoufie, Sari T. S.; Ahmed, Samrein B. M.; Shabana, Mona; Ahmed, Ahmed; Alqahtani, Saad S.; Alqahtani, Ali S.; Alqahtani, Ali M. et al. (21 May 2021). "Pathogenesis and Management of COVID-19". Journal of Xenobiotics 11 (2): 77–93. doi:10.3390/jox11020006. PMID 34063739.
- ↑ King, Andrew M. Q.; Adams, Michael J.; Carstens, Eric B. et al., eds. (2012-01-01), "Order - Nidovirales" (in en), Virus Taxonomy (Elsevier): 784–794, doi:10.1016/B978-0-12-384684-6.00066-5, ISBN 978-0-12-384684-6, http://www.sciencedirect.com/science/article/pii/B9780123846846000665, retrieved 2020-06-08
- ↑ Bukhari, Khulud; Mulley, Geraldine; Gulyaeva, Anastasia A.; Zhao, Lanying; Shu, Guocheng; Jiang, Jianping; Neuman, Benjamin W. (2018-11-01). "Description and initial characterization of metatranscriptomic nidovirus-like genomes from the proposed new family Abyssoviridae, and from a sister group to the Coronavirinae, the proposed genus Alphaletovirus" (in en). Virology 524: 160–171. doi:10.1016/j.virol.2018.08.010. ISSN 0042-6822. PMID 30199753.
- ↑ "The coronavirus nucleocapsid is a multifunctional protein". Viruses 6 (8): 2991–3018. August 2014. doi:10.3390/v6082991. PMID 25105276.
- ↑ Schoeman, Dewald; Fielding, Burtram C. (December 2019). "Coronavirus envelope protein: current knowledge". Virology Journal 16 (1): 69. doi:10.1186/s12985-019-1182-0. PMID 31133031.
- ↑ Jump up to: 6.0 6.1 Su S, Wong G, Shi W, Liu J, Lai ACK, Zhou J, Liu W, Bi Y, Gao GF. Epidemiology, Genetic Recombination, and Pathogenesis of Coronaviruses. Trends Microbiol. 2016 Jun;24(6):490-502. DOI: 10.1016/j.tim.2016.03.003. Epub 2016 Mar 21. Review. PMID 27012512
- ↑ "Virus Taxonomy: 2019 Release". International Committee on Taxonomy of Viruses. https://ictv.global/taxonomy.
- ↑ "The species Severe acute respiratory syndrome related coronavirus: classifying 2019-nCoV and naming it SARS-CoV-2". Nature Microbiology 5 (4): 536–44. 2020. doi:10.1038/s41564-020-0695-z. PMID 32123347.
- ↑ "Bat Coronaviruses in China". Viruses 11 (3): 210. March 2019. doi:10.3390/v11030210. PMID 30832341.
- ↑ "2017.012-015S" (in en) (xlsx). October 2018. https://ictv.global/ictv/proposals/2017.012_015S.A.v1.Nidovirales.zip.
- ↑ "The 2019–2020 Novel Coronavirus (Severe Acute Respiratory Syndrome Coronavirus 2) Pandemic: A Joint American College of Academic International Medicine‑World Academic Council of Emergency Medicine Multidisciplinary COVID‑19 Working Group Consensus Paper". https://www.researchgate.net/publication/340903626.
- ↑ "A planarian nidovirus expands the limits of RNA genome size". PLOS Pathogens 14 (11): e1007314. November 2018. doi:10.1371/journal.ppat.1007314. PMID 30383829.
External links
- Virus Pathogen Database and Analysis Resource (ViPR): Coronaviridae
- Archived Web page from 2006 on coronaviruses
- Focus on Coronaviruses (Microbiology Blog post from 2007)
- Viralzone: Coronaviridae
- International Committee on Taxonomy of Viruses
Wikidata ☰ Q1134583 entry
 |