Biology:Mobilome
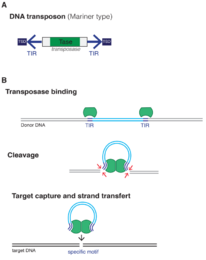
The mobilome is the entire set of mobile genetic elements in a genome. Mobilomes are found in eukaryotes,[1] prokaryotes,[2] and viruses.[3] The compositions of mobilomes differ among lineages of life, with transposable elements being the major mobile elements in eukaryotes, and plasmids and prophages being the major types in prokaryotes.[4] Virophages contribute to the viral mobilome.[5]
Mobilome in eukaryotes
Transposable elements are elements that can move about or propagate within the genome, and are the major constituents of the eukaryotic mobilome.[4] Transposable elements can be regarded as genetic parasites because they exploit the host cell's transcription and translation mechanisms to extract and insert themselves in different parts of the genome, regardless of the phenotypic effect on the host.[6]
Eukaryotic transposable elements were first discovered in maize (Zea mays) in which kernels showed a dotted color pattern.[7] Barbara McClintock described the maize Ac/Ds system in which the Ac locus promotes the excision of the Ds locus from the genome, and excised Ds elements can mutate genes responsible for pigment production by inserting into their coding regions.[8]
Other examples of transposable elements include: yeast (Saccharomyces cerevisiae) Ty elements, a retrotransposon which encodes a reverse transcriptase to convert its mRNA transcript into DNA which can then insert into other parts of the genome;[9][10] and fruit fly (Drosophila melanogaster) P-elements, which randomly inserts into the genome to cause mutations in germ line cells, but not in somatic cells.[11]
Mobilome in prokaryotes
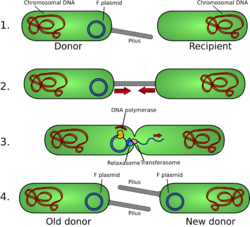
Plasmids were discovered in the 1940s as genetic materials outside of bacterial chromosomes.[12] Prophages are genomes of bacteriophages (a type of virus) that are inserted into bacterial chromosomes; prophages can then be spread to other bacteria through the lytic cycle and lysogenic cycle of viral replication.[13]
While transposable elements are also found in prokaryotic genomes,[14] the most common mobile genetic elements in the prokaryotic genome are plasmids and prophages.[4]
Plasmids and prophages can move between genomes through bacterial conjugation, allowing horizontal gene transfer.[15] Plasmids often carry genes that are responsible for bacterial antibiotic resistance; as these plasmids replicate and pass from one genome to another, the whole bacterial population can quickly adapt to the antibiotic.[16][17] Prophages can loop out of bacterial chromosomes to produce bacteriophages that go on to infect other bacteria with the prophages; this allows prophages to propagate quickly among the bacterial population, to the harm of the bacterial host.[13]
Mobilome in viruses
Discovered in 2008 in a strain of Acanthamoeba castellanii mimivirus,[18] virophages are an element of the virus mobilome.[5] Virophages are viruses that replicate only when host cells are co-infected with helper viruses.[19] Following co-infection, helper viruses exploit the host cell's transcription/translation machinery to produce their own machinery; virophages replicate through the machinery of either the host cell or the viruses.[19] The replication of virophages can negatively impact the replication of helper viruses.[18][20]
Sputnik[18][21] and mavirus[22] are examples of virophages.
References
- ↑ "The role of selfish genetic elements in eukaryotic evolution". Nature Reviews. Genetics 2 (8): 597–606. August 2001. doi:10.1038/35084545. PMID 11483984.
- ↑ "Mobile elements as a combination of functional modules". Plasmid 47 (1): 26–35. January 2002. doi:10.1006/plas.2001.1552. PMID 11798283.
- ↑ "A virus mutant with an insertion of a copia-like transposable element". Nature 299 (5883): 562–4. October 1982. doi:10.1038/299562a0. PMID 6289125. Bibcode: 1982Natur.299..562M.
- ↑ 4.0 4.1 4.2 Gogarten, Maria Boekels; Gogarten, Johann Peter; Olendzenski, Lorraine C., eds (2009). "Defining the Mobilome". Horizontal Gene Transfer: Genomes in Flux. Methods in Molecular Biology. 532. Humana Press. pp. 13–27. doi:10.1007/978-1-60327-853-9_2. ISBN 9781603278539.
- ↑ 5.0 5.1 "The Expanding Family of Virophages". Viruses 8 (11): 317. November 2016. doi:10.3390/v8110317. PMID 27886075.
- ↑ "Horizontal transposon transfer in eukarya: detection, bias, and perspectives". Genome Biology and Evolution 4 (8): 689–99. 2012. doi:10.1093/gbe/evs055. PMID 22798449.
- ↑ "The origins of maize genetics". Nature Reviews. Genetics 2 (11): 898–905. November 2001. doi:10.1038/35098524. PMID 11715045.
- ↑ "The origin and behavior of mutable loci in maize". Proceedings of the National Academy of Sciences of the United States of America 36 (6): 344–55. June 1950. doi:10.1073/pnas.36.6.344. PMID 15430309. Bibcode: 1950PNAS...36..344M.
- ↑ "Reverse transcriptase activity and Ty RNA are associated with virus-like particles in yeast". Nature 318 (6046): 583–6. December 1985. doi:10.1038/318583a0. PMID 2415827. Bibcode: 1985Natur.318..583M.
- ↑ "Ty element transposition: reverse transcriptase and virus-like particles". Cell 42 (2): 507–17. September 1985. doi:10.1016/0092-8674(85)90108-4. PMID 2411424.
- ↑ "Tissue specificity of Drosophila P element transposition is regulated at the level of mRNA splicing". Cell 44 (1): 7–19. January 1986. doi:10.1016/0092-8674(86)90480-0. PMID 3000622.
- ↑ "The cytoplasm in heredity". Heredity 4 (1): 11–36. April 1950. doi:10.1038/hdy.1950.2. PMID 15415003.
- ↑ 13.0 13.1 "Lysogenic versus lytic cycle of phage multiplication". Cold Spring Harbor Symposia on Quantitative Biology 18: 65–70. 1953-01-01. doi:10.1101/SQB.1953.018.01.014. PMID 13168970.
- ↑ "Nomenclature of transposable elements in prokaryotes". Gene 5 (3): 197–206. March 1979. doi:10.1016/0378-1119(79)90078-7. PMID 467979.
- ↑ "Horizontal gene transfer in human pathogens". Critical Reviews in Microbiology 41 (1): 101–8. February 2015. doi:10.3109/1040841X.2013.804031. PMID 23862575. https://www.repository.cam.ac.uk/bitstream/1810/244728/1/Juhas%20embargo%20text.pdf.
- ↑ "Plasmid-mediated horizontal gene transfer is a coevolutionary process". Trends in Microbiology 20 (6): 262–7. June 2012. doi:10.1016/j.tim.2012.04.003. PMID 22564249. http://eprints.whiterose.ac.uk/75385/1/Harrison_Brockhurst_Postprint.pdf.
- ↑ "Evolutionary consequences of antibiotic use for the resistome, mobilome and microbial pangenome". Frontiers in Microbiology 4: 4. 2013. doi:10.3389/fmicb.2013.00004. PMID 23386843.
- ↑ 18.0 18.1 18.2 "The virophage as a unique parasite of the giant mimivirus". Nature 455 (7209): 100–4. September 2008. doi:10.1038/nature07218. PMID 18690211. Bibcode: 2008Natur.455..100L.
- ↑ 19.0 19.1 "Mimivirus and its virophage". Annual Review of Genetics 43 (1): 49–66. 2009. doi:10.1146/annurev-genet-102108-134255. PMID 19653859.
- ↑ "Viva lavidaviruses! Five features of virophages that parasitize giant DNA viruses". PLOS Pathogens 15 (3): e1007592. March 2019. doi:10.1371/journal.ppat.1007592. PMID 30897185.
- ↑ "Structural studies of the Sputnik virophage". Journal of Virology 84 (2): 894–7. January 2010. doi:10.1128/JVI.01957-09. PMID 19889775.
- ↑ "Host genome integration and giant virus-induced reactivation of the virophage mavirus". Nature 540 (7632): 288–291. December 2016. doi:10.1038/nature20593. PMID 27929021. Bibcode: 2016Natur.540..288F. https://www.biorxiv.org/content/biorxiv/early/2016/10/18/068312.full.pdf.
 |
