Biology:Model lipid bilayer
A model lipid bilayer is any bilayer assembled in vitro, as opposed to the bilayer of natural cell membranes or covering various sub-cellular structures like the nucleus. They are used to study the fundamental properties of biological membranes in a simplified and well-controlled environment, and increasingly in bottom-up synthetic biology for the construction of artificial cells.[1] A model bilayer can be made with either synthetic or natural lipids. The simplest model systems contain only a single pure synthetic lipid. More physiologically relevant model bilayers can be made with mixtures of several synthetic or natural lipids.
There are many different types of model bilayers, each having experimental advantages and disadvantages. The first system developed was the black lipid membrane or “painted” bilayer, which allows simple electrical characterization of bilayers but is short-lived and can be difficult to work with. Supported bilayers are anchored to a solid substrate, increasing stability and allowing the use of characterization tools not possible in bulk solution. These advantages come at the cost of unwanted substrate interactions which can denature membrane proteins.
Black lipid membranes (BLM)
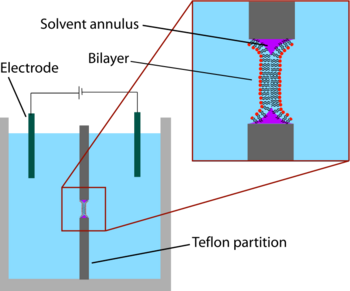
The earliest model bilayer system developed was the “painted” bilayer, also known as a “black lipid membrane.” The term “painted” refers to the process by which these bilayers are made. First, a small aperture is created in a thin layer of a hydrophobic material such as Teflon. Typically the diameter of this hole is a few tens of micrometers up to hundreds of micrometers. To form a BLM, the area around the aperture is first "pre-painted" with a solution of lipids dissolved in a hydrophobic solvent by applying this solution across the aperture with a brush, syringe, or glass applicator.[2] The solvent used must have a very high partition coefficient and must be relatively viscous to prevent immediate rupture. The most common solvent used is a mixture of decane and squalene.
After allowing the aperture to dry, salt solution (aqueous phase) is added to both sides of the chamber. The aperture is then "painted" with a lipid solution (generally the same solution that was used for pre-painting). A lipid monolayer spontaneously forms at the interface between the organic and aqueous phases on either side of the lipid/solvent droplet. Because the walls of the aperture are hydrophobic the lipid/solvent solution wets this interface, thinning the droplet in the center. Once the two sides of the droplet come close enough together, the lipid monolayers fuse, rapidly excluding the small remaining volume of solution. At this point a bilayer is formed in the center of the aperture, but a significant annulus of solvent remains at the perimeter. This annulus is required to maintain stability by acting as a bridge between the ~5 nm bilayer and the tens of micrometers thick sheet in which the aperture is made.[3]
The term “black” bilayer refers to the fact that they are dark in reflected light because the thickness of the membrane is only a few nanometers, so light reflecting off the back face destructively interferes with light reflecting off the front face. Indeed, this was one of the first clues that this technique produced a membrane of molecular-scale thickness.[4] Black lipid membranes are also well suited to electrical characterization because the two chambers separated by the bilayer are both accessible, allowing simple placement of large electrodes. For this reason, electrical characterization is one of the most important methods used in conjunction with painted lipid bilayers. Simple measurements indicate when a bilayer forms and when it breaks, as an intact bilayer has a large resistance (>GΩ) and a large capacitance (~2 µF/cm2). More advanced electrical characterization has been particularly important in the study of voltage gated ion channels. Membrane proteins such as ion channels typically cannot be incorporated directly into the painted bilayer during formation because immersion in an organic solvent would denature the protein. Instead, the protein is solubilized with a detergent and added to the aqueous solution after the bilayer is formed. The detergent coating allows these proteins to spontaneously insert into the bilayer over a period of minutes. Additionally, initial experiments have been performed which combine electrophysiological and structural investigations of black lipid membranes.[5] In another variation of the BLM technique, termed the bilayer punch, a glass pipet (inner diameter ~10-40 µm) is used as the electrode on one side of the bilayer in order to isolate a small patch of membrane.[6][7] This modification of the patch clamp technique enables low noise recording, even at high potentials (up to 600 mV), at the expense of additional preparation time.
The main problems associated with painted bilayers are residual solvent and limited lifetime. Some researchers believe that pockets of solvent trapped between the two bilayer leaflets can disrupt normal protein function. To overcome this limitation, Montal and Mueller developed a modified deposition technique that eliminates the use of a heavy non-volatile solvent. In this method, the aperture starts out above the water surface, completely separating the two fluid chambers. On the surface of each chamber, a monolayer is formed by applying lipids in a volatile solvent such as chloroform and waiting for the solvent to evaporate. The aperture is then lowered through the air-water interface and the two monolayers from the separate chambers are folded down against each other, forming a bilayer across the aperture.[8] The stability issue has proven more difficult to solve. Typically, a black lipid membrane will survive for less than an hour, precluding long-term experiments. This lifetime can be extended by precisely structuring the supporting aperture,[9] chemically crosslinking the lipids or gelling the surrounding solution to mechanically support the bilayer.[10] Work is ongoing in this area and lifetimes of several hours will become feasible.
Supported lipid bilayers (SLB)

Unlike a vesicle or a cell membrane in which the lipid bilayer is rolled into an enclosed shell, a supported bilayer is a planar structure sitting on a solid support. Because of this, only the upper face of the bilayer is exposed to free solution. This layout has advantages and drawbacks related to the study of lipid bilayers. One of the greatest advantages of the supported bilayer is its stability. SLBs will remain largely intact even when subject to high flow rates or vibration and, unlike black lipid membranes, the presence of holes will not destroy the entire bilayer. Because of this stability, experiments lasting weeks and even months are possible with supported bilayers while BLM experiments are usually limited to hours.[11] Another advantage of the supported bilayer is that, because it is on a flat hard surface, it is amenable to a number of characterization tools which would be impossible or would offer lower resolution if performed on a freely floating sample.
One of the clearest examples of this advantage is the use of mechanical probing techniques which require a direct physical interaction with the sample. Atomic force microscopy (AFM) has been used to image lipid phase separation,[12] formation of transmembrane nanopores followed by single protein molecule adsorption,[13] and protein assembly[14] with sub-nm accuracy without the need for a labeling dye. More recently, AFM has also been used to directly probe the mechanical properties of single bilayers[15] and to perform force spectroscopy on individual membrane proteins.[16] These studies would be difficult or impossible without the use of supported bilayers since the surface of a cell or vesicle is relatively soft and would drift and fluctuate over time. Another example of a physical probe is the use of the quartz crystal microbalance (QCM) to study binding kinetics at the bilayer surface.[17] Dual polarisation interferometry is a high resolution optical tool for characterising the order and disruption in lipid bilayers during interactions or phase transitions providing complementary data to QCM measurements.[18]
Many modern fluorescence microscopy techniques also require a rigidly-supported planar surface. Evanescent field methods such as total internal reflection fluorescence microscopy (TIRF) and surface plasmon resonance (SPR) can offer extremely sensitive measurement of analyte binding and bilayer optical properties but can only function when the sample is supported on specialized optically functional materials. Another class of methods applicable only to supported bilayers is those based on optical interference such as fluorescence interference contrast microscopy (FLIC) and reflection interference contrast microscopy (RICM) or interferometric scattering microscopy (iSCAT).[19][20] When the bilayer is supported on top of a reflective surface, variations in intensity due to destructive interference from this interface can be used to calculate with angstrom accuracy the position of fluorophores within the bilayer.[21] Both evanescent and interference techniques offer sub-wavelength resolution in only one dimension (z, or vertical). In many cases, this resolution is all that is needed. After all, bilayers are very small only in one dimension. Laterally, a bilayer can extend for many micrometres or even millimeters. But certain phenomena like dynamic phase rearrangement do occur in bilayers on a lateral sub-micrometre length scale. A promising approach to studying these structures is near field scanning optical microscopy (NSOM).[22] Like AFM, NSOM relies on the scanning of a micromachined tip to give a highly localized signal. But unlike AFM, NSOM uses an optical rather than physical interaction with the sample, potentially perturbing delicate structures to a lesser extent.

Another important capability of supported bilayers is the ability to pattern the surface to produce multiple isolated regions on the same substrate. This phenomenon was first demonstrated using scratches or metallic “corrals” to prevent mixing between adjacent regions while still allowing free diffusion within any one region.[23][24] Later work extended this concept by integrating microfluidics to demonstrate that stable composition gradients could be formed in bilayers,[25] potentially allowing massively parallel studies of phase segregation, molecular binding and cellular response to artificial lipid membranes. Creative utilization of the corral concept has also allowed studies of the dynamic reorganization of membrane proteins at the synaptic interface.[26]
One of the primary limitations of supported bilayers is the possibility of unwanted interactions with the substrate. Although supported bilayers generally do not directly touch the substrate surface, they are separated by only a very thin water gap. The size and nature of this gap depends on the substrate material[27] and lipid species but is generally about 1 nm for zwitterionic lipids supported on silica, the most common experimental system.[28][29] Because this layer is so thin there is extensive hydrodynamic coupling between the bilayer and the substrate, resulting in a lower diffusion coefficient in supported bilayers than for free bilayers of the same composition.[30] A certain percentage of the supported bilayer will also be completely immobile, although the exact nature of and reason for these “pinned” sites is still uncertain. For high quality liquid phase supported bilayers the immobile fraction is typically around 1-5%. To quantify the diffusion coefficient and mobile fraction, researchers studying supported bilayers will often report FRAP data.
Unwanted substrate interactions are a much greater problem when incorporating integral membrane proteins, particularly those with large domains sticking out beyond the core of the bilayer. Because the gap between bilayer and substrate is so thin these proteins will often become denatured on the substrate surface and therefore lose all functionality.[31] One approach to circumvent this problem is the use of polymer tethered bilayers. In these systems the bilayer is supported on a loose network of hydrated polymers or hydrogel which acts as a spacer and theoretically prevents denaturing substrate interactions.[32] In practice, some percentage of the proteins will still lose mobility and functionality, probably due to interactions with the polymer/lipid anchors.[30] Research in this area is ongoing.
Tethered bilayer lipid membranes (t-BLM)
The use of a tethered bilayer lipid membrane (t-BLM) further increases the stability of supported membranes by chemically anchoring the lipids to the solid substrate.[33]
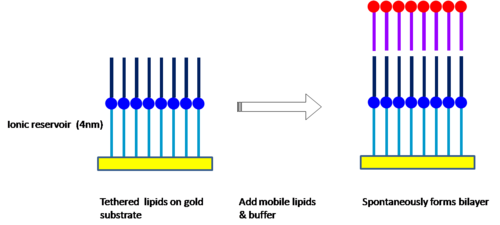
Gold can be used as a substrate because of its inert chemistry and thiolipids for covalent binding to the gold. Thiolipids are composed of lipid derivatives, extended at their polar head-groups by hydrophilic spacers which terminate in a thiol or disulphide group that forms a covalent bond with gold, forming self assembled monolayers (SAM).
The limitation of the intra-membrane mobility of supported lipid bilayers can be overcome by introducing half-membrane spanning tether lipids[34] with benzyl disulphide (DPL) and synthetic archaea analogue full membrane spanning lipids with phytanoly chains to stabilize the structure and polyethyleneglycol units as a hydrophilic spacer. Bilayer formation is achieved by exposure of the lipid coated gold substrate to outer layer lipids either in an ethanol solution or in liposomes.[35]
The advantage of this approach is that because of the hydrophilic space of around 4 nm, the interaction with the substrate is minimal and the extra space allows the introduction of protein ion channels into the bilayer. Additionally the spacer layer creates an ionic reservoir[36] that readily enables ac electrical impedance measurement across the bilayer.
Vesicles
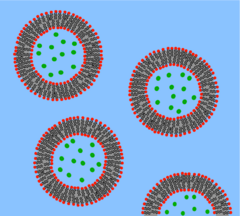
A vesicle is a lipid bilayer rolled up into a spherical shell, enclosing a small amount of water and separating it from the water outside the vesicle. Because of this fundamental similarity to the cell membrane, vesicles have been used extensively to study the properties of lipid bilayers. Another reason vesicles have been used so frequently is that they are relatively easy to make. If a sample of dehydrated lipid is exposed to water it will spontaneously form vesicles.[37] These initial vesicles are typically multilamellar (many-walled) and are of a wide range of sizes from tens of nanometers to several micrometres.[38] Methods such as sonication or extrusion through a membrane are needed to break these initial vesicles into smaller, single-walled vesicles of uniform diameter known as small unilamellar vesicles (SUVs). SUVs typically have diameters between 50 and 200 nm.[39] Alternatively, rather than synthesizing vesicles it is possible to simply isolate them from cell cultures or tissue samples.[40] Vesicles are used to transport lipids, proteins and many other molecules within the cell as well as into or out of the cell. These naturally isolated vesicles are composed of a complex mixture of different lipids and proteins so, although they offer greater realism for studying specific biological phenomena, simple artificial vesicles are preferred for studies of fundamental lipid properties.
Since artificial SUVs can be made in large quantities they are suitable for bulk material studies such as x-ray diffraction to determine lattice spacing[41] and differential scanning calorimetry to determine phase transitions.[42] Dual polarisation interferometry can measure unilamelar and multilamelar structures and insertion into and disruption of the vesicles in a label free assay format.[43] Vesicles can also be labeled with fluorescent dyes to allow sensitive FRET-based fusion assays.[44]
In spite of the fluorescent labeling, it is often difficult to perform detailed imaging on SUVs simply because they are so small. To combat this problem, researchers use giant unilamellar vesicles (GUVs). GUVs are large enough (1 - 200 µm) to be studied using traditional fluorescence microscopy and are within the same size range as most biological cells. Thus, they are used as mimicries of cell membranes for in vitro studies in molecular and cell biology. Many of the studies of lipid rafts in artificial lipid systems have been performed with GUVs for this reason.[45] Compared to supported bilayers, GUVs present a more “natural” environment since there is no rigid surface that might induce defects, affect the properties of the membrane or denature proteins. Therefore, GUVs are frequently used to study membrane-remodeling and other protein-membrane interactions in vitro. A variety of methods exist to encapsulate proteins or other biological reactants within such vesicles, making GUVs an ideal system for the in vitro recreation (and investigation) of cell functions in cell-like model membrane environments.[46] These methods include microfluidic methods, which allow for a high-yield production of vesicles with consistent sizes.[47]
Droplet Interface Bilayers
Droplet Interface Bilayers (DIBs) are phospholipid-encased droplets that form bilayers when they are put into contact.[48][49] The droplets are surrounded by oil and phospholipids are dispersed in either the water or oil.[48] As a result, the phospholipids spontaneously form a monolayer at each of the oil-water interfaces.[48] DIBs can be formed to create tissue-like material with the ability to form asymmetric bilayers, reconstitute proteins and protein channels or made for use in studying electrophysiology.[50][51][52][53][54] Extended DIB networks can be formed either by employing droplet microfluidic devices or using droplet printers.[54][55]
Micelles, bicelles and nanodiscs
Detergent micelles[56] are another class of model membranes that are commonly used to purify and study membrane proteins, although they lack a lipid bilayer. In aqueous solutions, micelles are assemblies of amphipathic molecules with their hydrophilic heads exposed to solvent and their hydrophobic tails in the center. Micelles can solubilize membrane proteins by partially encapsulating them and shielding their hydrophobic surfaces from solvent.
Bicelles are a related class of model membrane,[57] typically made of two lipids, one of which forms a lipid bilayer while the other forms an amphipathic, micelle-like assembly shielding the bilayer center from surrounding solvent molecules. Bicelles can be thought of as a segment of bilayer encapsulated and solubilized by a micelle. Bicelles are much smaller than liposomes, and so can be used in experiments such as NMR spectroscopy where the larger vesicles are not an option.
Nanodiscs [58] consist of a segment of bilayer encapsulated by an amphipathic protein coat, rather than a lipid or detergent layer. Nanodiscs are more stable than bicelles and micelles at low concentrations, and are very well-defined in size (depending on the type of protein coat, between 10 and 20 nm). Membrane proteins incorporated into and solubilized by Nanodiscs can be studied by a wide variety of biophysical techniques.[59][60]
References
- ↑ "Artificial cell mimics as simplified models for the study of cell biology". Experimental Biology and Medicine 242 (13): 1309–1317. July 2017. doi:10.1177/1535370217711441. PMID 28580796.
- ↑ "Reconstitution of cell membrane structure in vitro and its transformation into an excitable system". Nature 194 (4832): 979–80. June 1962. doi:10.1038/194979a0. PMID 14476933. Bibcode: 1962Natur.194..979M.
- ↑ "Analysis of the torus surrounding planar lipid bilayer membranes". Biophysical Journal 12 (4): 432–45. April 1972. doi:10.1016/s0006-3495(72)86095-8. PMID 5019479. Bibcode: 1972BpJ....12..432W.
- ↑ "Formation of "black" lipid membranes by oxidation products of cholesterol". Nature 212 (5063): 718–719. 1966. doi:10.1038/212718a0. Bibcode: 1966Natur.212..718T.
- ↑ "Hard X-Ray Phase Contrast Imaging of Black lipid membranes". Applied Physics Letters 95 (20): 203703. November 2009. doi:10.1063/1.3263946. Bibcode: 2009ApPhL..95t3703B.
- ↑ "Ion movement through gramicidin A channels. Single-channel measurements at very high potentials". Biophysical Journal 41 (2): 119–33. February 1983. doi:10.1016/S0006-3495(83)84414-2. PMID 6188500. Bibcode: 1983BpJ....41..119A.
- ↑ "Single molecule methods for monitoring changes in bilayer elastic properties". Journal of Visualized Experiments 21 (21): e1032. November 2008. doi:10.3791/1032. PMID 19066527.
- ↑ "Formation of bimolecular membranes from lipid monolayers and a study of their electrical properties". Proceedings of the National Academy of Sciences of the United States of America 69 (12): 3561–6. December 1972. doi:10.1073/pnas.69.12.3561. PMID 4509315. Bibcode: 1972PNAS...69.3561M.
- ↑ "X-ray structure analysis of free-standing lipid membranes facilitated by micromachined apertures". Langmuir: The ACS Journal of Surfaces and Colloids 24 (9): 4952–8. May 2008. doi:10.1021/la703704x. PMID 18370435.
- ↑ "Long‐Lived Planar Lipid Bilayer Membranes Anchored to an In Situ Polymerized Hydrogel.". Advanced Materials 20 (1): 84–9. January 2008. doi:10.1002/adma.200700810.
- ↑ "Deposition of highly resistive lipid bilayer on silicon-silicon dioxide electrode and incorporation of gramicidin studied by ac impedance spectroscopy". Electrochimica Acta 47 (5): 791–798. 2001. doi:10.1016/s0013-4686(01)00759-9.
- ↑ "Lipid asymmetry in DLPC/DSPC-supported lipid bilayers: a combined AFM and fluorescence microscopy study". Biophysical Journal 90 (1): 228–37. January 2006. doi:10.1529/biophysj.105.067066. PMID 16214871. Bibcode: 2006BpJ....90..228L.
- ↑ "Interaction of nanoparticles with lipid membrane". Nano Letters 8 (3): 941–4. March 2008. doi:10.1021/nl080080l. PMID 18254602. Bibcode: 2008NanoL...8..941R.
- ↑ "Observing single biomolecules at work with the atomic force microscope". Nature Structural Biology 7 (9): 715–8. September 2000. doi:10.1038/78929. PMID 10966636.
- ↑ "Mechanical properties of pore-spanning lipid bilayers probed by atomic force microscopy". Biophysical Journal 91 (1): 217–26. July 2006. doi:10.1529/biophysj.106.081398. PMID 16617084. Bibcode: 2006BpJ....91..217S.
- ↑ "Unfolding pathways of individual bacteriorhodopsins". Science 288 (5463): 143–6. April 2000. doi:10.1126/science.288.5463.143. PMID 10753119. Bibcode: 2000Sci...288..143O.
- ↑ "A kinetic study of concanavalin A binding to glycolipid monolayers by using a quartz-crystal microbalance.". Journal of the American Chemical Society 116 (25): 11209–12. December 1994. doi:10.1021/ja00104a001.
- ↑ "Optical anisotropy of supported lipid structures probed by waveguide spectroscopy and its application to study of supported lipid bilayer formation kinetics". Analytical Chemistry 80 (10): 3666–76. May 2008. doi:10.1021/ac800027s. PMID 18422336.
- ↑ "Direct observation and control of supported lipid bilayer formation with interferometric scattering microscopy". ACS Nano 7 (12): 10662–70. December 2013. doi:10.1021/nn403367c. PMID 24251388. https://ora.ox.ac.uk/objects/uuid:013ec465-bf31-4298-bc40-60142ebcc631.
- ↑ "Dynamic label-free imaging of lipid nanodomains". Proceedings of the National Academy of Sciences of the United States of America 112 (40): 12299–303. October 2015. doi:10.1073/pnas.1508483112. PMID 26401022. Bibcode: 2015PNAS..11212299D.
- ↑ "Measuring lipid asymmetry in planar supported bilayers by fluorescence interference contrast microscopy". Langmuir: The ACS Journal of Surfaces and Colloids 21 (4): 1377–88. February 2005. doi:10.1021/la047654w. PMID 15697284.
- ↑ "Submicron structure in L-alpha-dipalmitoylphosphatidylcholine monolayers and bilayers probed with confocal, atomic force, and near-field microscopy". Biophysical Journal 75 (1): 342–53. July 1998. doi:10.1016/s0006-3495(98)77518-6. PMID 9649391. Bibcode: 1998BpJ....75..342H.
- ↑ "Micropatterning fluid lipid bilayers on solid supports". Science (New York, N.Y.) 275 (5300): 651–3. January 1997. doi:10.1126/science.275.5300.651. PMID 9005848.
- ↑ "Substrate−Membrane Interactions: Mechanisms for Imposing Patterns on a Fluid Bilayer Membrane". Langmuir 14 (12): 3347–50. 1998. doi:10.1021/la9711701.
- ↑ "Spatially Selective Manipulation of Supported Lipid Bilayers by Laminar Flow: Steps Toward Biomembrane Microfluidics". Langmuir 19 (5): 1624–1631. 2003. doi:10.1021/la0263413.
- ↑ "Nonequilibrium Adhesion Patterns at Lipid Bilayer Junctions". Journal of Physical Chemistry B 108 (2): 649–57. 2004. doi:10.1021/jp035543k.
- ↑ "Formation and characterization of fluid lipid bilayers on alumina". Langmuir 24 (22): 12734–7. November 2008. doi:10.1021/la802726u. PMID 18942863.
- ↑ "Neutron reflectivity and atomic force microscopy studies of a lipid bilayer in water adsorbed to the surface of a silicon single crystal". Langmuir 12 (5): 1343–1350. 1996. doi:10.1021/la950580r.
- ↑ "Structure of an adsorbed dimyristoylphosphatidylcholine bilayer measured with specular reflection of neutrons". Biophysical Journal 59 (2): 289–94. February 1991. doi:10.1016/S0006-3495(91)82222-6. PMID 2009353. Bibcode: 1991BpJ....59..289J.
- ↑ 30.0 30.1 "Lipid mono- and bilayer supported on polymer films: composite polymer-lipid films on solid substrates". Biophysical Journal 67 (1): 217–26. July 1994. doi:10.1016/s0006-3495(94)80472-2. PMID 7918990. Bibcode: 1994BpJ....67..217K.
- ↑ "Solid supported lipid bilayers: From biophysical studies to sensor design". Surface Science Reports 61 (10): 429–444. November 2006. doi:10.1016/j.surfrep.2006.06.001. PMID 32287559. Bibcode: 2006SurSR..61..429C.
- ↑ "Polymer-cushioned bilayers. II. An investigation of interaction forces and fusion using the surface forces apparatus". Biophysical Journal 77 (3): 1458–68. September 1999. doi:10.1016/s0006-3495(99)76993-6. PMID 10465756. Bibcode: 1999BpJ....77.1458W.
- ↑ "Incorporation of Membrane Proteins in Solid-Supported Lipid Layers". Angew. Chem. 34 (18): 2056–2058. 1995. doi:10.1002/anie.199520561.
- ↑ "A biosensor that uses ion-channel switches". Nature 387 (6633): 580–3. June 1997. doi:10.1038/42432. PMID 9177344. Bibcode: 1997Natur.387..580C.
- ↑ "A new class of thiolipids for the attachment of lipid bilayers on gold surfaces". Langmuir 10: 197–210. 1994. doi:10.1021/la00013a029.
- ↑ "Tethered-bilayer lipid membranes as a support for membrane-active peptides". Biochemical Society Transactions 29 (Pt 4): 613–7. August 2001. doi:10.1042/BST0290613. PMID 11498038.
- ↑ "Negative staining of phospholipids and their structural modification by surface-active agents as observed in the electron microscope.". Journal of Molecular Biology 8 (5): 660–668. January 1964. doi:10.1016/S0022-2836(64)80115-7. PMID 14187392.
- ↑ "The mechanism of vesicle formation". The Biochemical Journal 256 (1): 1–11. November 1988. doi:10.1042/bj2560001. PMID 3066342.
- ↑ F Szoka and D Papahadjopoulos."Comparative Properties and Methods of Preparation of Lipid Vesicles (Liposomes)." Annual Review of Biophysics and Bioengineering. 9. (1980) 467-508.
- ↑ "VAMP-1: a synaptic vesicle-associated integral membrane protein". Proceedings of the National Academy of Sciences of the United States of America 85 (12): 4538–42. June 1988. doi:10.1073/pnas.85.12.4538. PMID 3380805. Bibcode: 1988PNAS...85.4538T.
- ↑ "Phospholipid model membranes. I. Structural characteristics of hydrated liquid crystals". Biochimica et Biophysica Acta (BBA) - Biomembranes 135 (4): 624–38. September 1967. doi:10.1016/0005-2736(67)90094-6. PMID 4167394.
- ↑ "The volume change in lipid bilayer lamellae at the crystalline-liquid crystalline phase transition.". Chemistry and Physics of Lipids 7 (4): 324–35. December 1971. doi:10.1016/0009-3084(71)90010-7.
- ↑ "Quantifying the effects of melittin on liposomes". Biochimica et Biophysica Acta (BBA) - Biomembranes 1768 (1): 13–20. January 2007. doi:10.1016/j.bbamem.2006.05.016. PMID 17092481.
- ↑ "Lipid bilayer vesicle fusion: intermediates captured by high-speed microfluorescence spectroscopy". Biophysical Journal 85 (3): 1585–99. September 2003. doi:10.1016/S0006-3495(03)74590-1. PMID 12944275. Bibcode: 2003BpJ....85.1585L.
- ↑ "Lipid rafts reconstituted in model membranes". Biophysical Journal 80 (3): 1417–28. March 2001. doi:10.1016/S0006-3495(01)76114-0. PMID 11222302. Bibcode: 2001BpJ....80.1417D.
- ↑ "Protein Reconstitution Inside Giant Unilamellar Vesicles". Annual Review of Biophysics 50: 525–548. March 2021. doi:10.1146/annurev-biophys-100620-114132. PMID 33667121.
- ↑ "Stepwise synthesis of giant unilamellar vesicles on a microfluidic assembly line". Journal of the American Chemical Society 133 (9): 2798–800. March 2011. doi:10.1021/ja109137s. PMID 21309555.
- ↑ 48.0 48.1 48.2 "Droplet interface bilayers". Molecular BioSystems 4 (12): 1191–208. December 2008. doi:10.1039/b808893d. PMID 19396383.
- ↑ "Lipid bilayer formation by contacting monolayers in a microfluidic device for membrane protein analysis". Analytical Chemistry 78 (24): 8169–74. December 2006. doi:10.1021/ac0613479. PMID 17165804.
- ↑ "Asymmetric droplet interface bilayers". Journal of the American Chemical Society 130 (18): 5878–9. May 2008. doi:10.1021/ja802089s. PMID 18407631.
- ↑ "In vitro reconstitution of eukaryotic ion channels using droplet interface bilayers". Journal of the American Chemical Society 133 (24): 9370–5. June 2011. doi:10.1021/ja200128n. PMID 21591742.
- ↑ "Activation of bacterial channel MscL in mechanically stimulated droplet interface bilayers". Scientific Reports 5: 13726. September 2015. doi:10.1038/srep13726. PMID 26348441. Bibcode: 2015NatSR...513726N.
- ↑ "Determining membrane capacitance by dynamic control of droplet interface bilayer area". Langmuir 27 (23): 14335–42. December 2011. doi:10.1021/la203081v. PMID 21978255.
- ↑ 54.0 54.1 "A tissue-like printed material". Science 340 (6128): 48–52. April 2013. doi:10.1126/science.1229495. PMID 23559243. Bibcode: 2013Sci...340...48V.
- ↑ "Light-Patterned Current Generation in a Droplet Bilayer Array" (in En). Scientific Reports 7: 46585. April 2017. doi:10.1038/srep46585. PMID 28417964. Bibcode: 2017NatSR...746585R.
- ↑ "Membrane proteins, lipids and detergents: not just a soap opera". Biochimica et Biophysica Acta (BBA) - Biomembranes 1666 (1–2): 105–17. November 2004. doi:10.1016/j.bbamem.2004.04.011. PMID 15519311.
- ↑ "Improved low pH bicelle system for orienting macromolecules over a wide temperature range". Journal of Biomolecular NMR 13 (4): 387–91. April 1999. doi:10.1023/a:1008360022444. PMID 10353198.
- ↑ "Reconstitution of Membrane Proteins in Phospholipid Bilayer Nanodiscs". Chapter 11 - Reconstitution of membrane proteins in phospholipid bilayer nanodiscs. Methods in Enzymology. 464. 2009. pp. 211–31. doi:10.1016/s0076-6879(09)64011-8. ISBN 9780123749697.
- ↑ "High-Level Cell-Free Production of Membrane Proteins with Nanodiscs". Cell-Free Protein Synthesis. Methods in Molecular Biology. 1118. 2014. pp. 109–30. doi:10.1007/978-1-62703-782-2_7. ISBN 978-1-62703-781-5.
- ↑ "Co-translational association of cell-free expressed membrane proteins with supplied lipid bilayers". Molecular Membrane Biology 30 (1): 75–89. February 2013. doi:10.3109/09687688.2012.693212. PMID 22716775.
 |
