Biology:Nucleomorph
This article is missing information about inclusions including nucleolus (plus rDNA!), division behavior, mentioned in refs of doi:10.1007/s00709-017-1153-5 (RG link). (October 2021) |
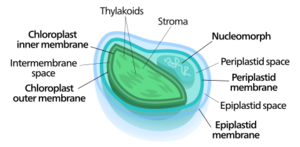
Nucleomorphs are small, vestigial eukaryotic nuclei found between the inner and outer pairs of membranes in certain plastids. They are thought to be vestiges of primitive red and green algal nuclei that were engulfed by a larger eukaryote. Because the nucleomorph lies between two sets of membranes, nucleomorphs support the endosymbiotic theory and are evidence that the plastids containing them are complex plastids. Having two sets of membranes indicate that the plastid, a prokaryote, was engulfed by a eukaryote, an alga, which was then engulfed by another eukaryote, the host cell, making the plastid an example of secondary endosymbiosis.[1][2]
Organisms with known nucleomorphs
So far, only two monophyletic groups of organisms are known to contain plastids with a vestigial nucleus or nucleomorph: the cryptomonads[3] of the supergroup Chromista and the chlorarachniophytes[4] of the supergroup Rhizaria, both of which have examples of sequenced nucleomorph genomes.[3][4] Studies of the genomic organization and of the molecular phylogeny have shown that the nucleomorph of the cryptomonads used to be the nucleus of a red alga, whereas the nucleomorph of the chlorarchniophytes was the nucleus of a green alga. In both groups of organisms the plastids originate from engulfed photoautotrophic eukaryotes.
Of the two known plastids that contain nucleomorphs, both have four membranes, the nucleomorph residing in the periplastidial compartment, evidence of being engulfed by a eukaryote through phagocytosis.[1]
In addition, some species within the dinoflagellates that have gone through tertiary endosymbiosis also have endosymbionts with both a nucleus and mitochondria present.[5]
Nucleomorph genome
Nucleomorphs represent some of the smallest genomes ever sequenced. After the red or green alga was engulfed by a cryptomonad or chlorarachniophyte, respectively, its genome was reduced. The nucleomorph genomes of both cryptomonads and chlorarachniophytes converged upon a similar size from larger genomes. They retained only three chromosomes and many genes were transferred to the nucleus of the host cell, while others were lost entirely.[1] Chlorarachniophytes contain a nucleomorph genome that is diploid and cryptomonads contain a nucleomorph genome that is tetraploid.[6] The unique combination of host cell and complex plastid results in cells with four genomes: two prokaryotic genomes (mitochondrion and plastid of the red or green algae) and two eukaryotic genomes (nucleus of host cell and nucleomorph).
The model cryptomonad Guillardia theta became an important focus for scientists studying nucleomorphs. Its complete nucleomorph sequence was published in 2001, coming in at 551 Kbp. The G. theta sequence gave insight as to what genes were retained in nucleomorphs. Most of the genes that moved to the host cell involved protein synthesis, leaving behind a compact genome with mostly single-copy “housekeeping” genes (affecting transcription, translation, protein folding and degradation and splicing) and no mobile elements. The genome contains 513 genes, 465 of which code for protein. Thirty genes are considered “plastid” genes, coding for plastid proteins.[1][7]
The genome sequence of another organism, the chlorarachniophyte Bigelowiella natans indicates that its nucleomorph is probably the vestigial nucleus of a green alga, whereas the nucleomorph in G. theta probably came from a red alga. The B. natans genome is smaller than that of G. theta, with about 373 Kbp and contains 293 protein-coding genes as compared to the 465 genes in G. theta. B. natans also only has 17 genes that code for plastid proteins, again fewer than G. theta. Comparisons between the two organisms have shown that B. natans contains significantly more introns (852) than G. theta (17). B. natans also had smaller introns, ranging from 18-21 bp, whereas G. theta’s introns ranged from 42-52 bp.[1]
Both the genomes of B. natans and G. theta display evidence of genome reduction besides elimination of genes and tiny size, including elevated composition of adenine (A) and thymine (T), and high substitution rates.[4][7][8]
Persistence of nucleomorphs
There are no recorded instances of vestigial nuclei in any other secondary plastid-containing organisms, yet they have been retained independently in the cryptomonads and chlorarachniophytes. Plastid gene transfer happens frequently in many organisms, and it is unusual that these nucleomorphs have not disappeared entirely. One theory as to why these nucleomorphs have not disappeared as they have in other groups is that introns present in nucleomorphs are not recognized by host spliceosomes because they are too small and therefore cannot be cut and later incorporated into host DNA.
Nucleomorphs also often code for many of their own critical functions, like transcription and translation.[9] Some say that as long as there exists a gene in the nucleomorph that codes for proteins necessary for the plastid’s functioning that are not produced by the host cell, the nucleomorph will persist.[1] In cryptophytes and chlorarachniophytes all DNA transfer between the nucleomorph and host genome seems to have ceased, but the process is still going on in a few dinoflagellates.[10]
See also
References
- ↑ 1.0 1.1 1.2 1.3 1.4 1.5 Archibald, J.M.; Lane, C.E. (2009). "Going, Going, Not Quite Gone: Nucleomorphs as a Case Study in Nuclear Genome Reduction". Journal of Heredity 100 (5): 582–90. doi:10.1093/jhered/esp055. PMID 19617523.
- ↑ Reyes-Prieto, Adrian; Weber, Andreas P.M.; Bhattacharya, Debashish (2007). "The Origin and Establishment of the Plastid in Algae and Plants". Annual Review of Genetics 41 (1): 147–68. doi:10.1146/annurev.genet.41.110306.130134. PMID 17600460.
- ↑ 3.0 3.1 Lane, C. E.; Van Den Heuvel, K.; Kozera, C.; Curtis, B. A.; Parsons, B. J.; Bowman, S.; Archibald, J. M. (2007). "Nucleomorph genome of Hemiselmis andersenii reveals complete intron loss and compaction as a driver of protein structure and function". Proceedings of the National Academy of Sciences 104 (50): 19908–19913. doi:10.1073/pnas.0707419104. PMID 18077423. Bibcode: 2007PNAS..10419908L.
- ↑ 4.0 4.1 4.2 Gilson, P. R.; Su, V.; Slamovits, C. H.; Reith, M. E.; Keeling, P. J.; McFadden, G. I. (2006). "Complete nucleotide sequence of the chlorarachniophyte nucleomorph: Nature's smallest nucleus". Proceedings of the National Academy of Sciences 103 (25): 9566–9571. doi:10.1073/pnas.0600707103. PMID 16760254. Bibcode: 2006PNAS..103.9566G.
- ↑ Tertiary Endosymbiosis in Two Dinotoms Has Generated Little Change in the Mitochondrial Genomes of Their Dinoflagellate Hosts and Diatom Endosymbionts - PLOS
- ↑ Hirakawa, Yoshihisa; Ishida, Ken-Ichiro (2014-04-01). "Polyploidy of Endosymbiotically Derived Genomes in Complex Algae". Genome Biology and Evolution 6 (4): 974–980. doi:10.1093/gbe/evu071. ISSN 1759-6653. PMID 24709562.
- ↑ 7.0 7.1 Archibald, John M (2007). "Nucleomorph Genomes: Structure, Function, Origin and Evolution". BioEssays 29 (4): 392–402. doi:10.1002/bies.20551. PMID 17373660.
- ↑ Douglas, SEExpression error: Unrecognized word "etal". (2001). "The highly reduced genome of an enslaved algal nucleus". Nature 410 (6832): 1091–1096. doi:10.1038/35074092. PMID 11323671. Bibcode: 2001Natur.410.1091D.
- ↑ Curtis, Bruce et al. "Algal genomes reveal evolutionary mosaicism and the fate of nucleomorphs." Nature 492 :59-65
- ↑ Organellogenesis still a work in progress in novel dinoflagellates
External links
- Insight into the Diversity and Evolution of the Cryptomonad Nucleomorph Genome
- Cryptophyta at NCBI taxbrowser
- Cercozoa at NCBI taxbrowser
According to GenBank release 164 (Feb 2008), there are 13 Cercozoa and 181 Cryptophyta entries (an entry is the submission of a sequence to the DDBJ/EMBL/GenBank public database of sequences). Most sequenced organisms were:
Guillardia theta: 54; Rhodomonas salina: 18; Cryptomonas sp.: 15; Chlorarachniophyceae sp.:10; Cryptomonas paramecium: 9; Cryptomonas erosa: 7.
Note that the taxonomy used in the first section is probably outdated[verification needed]. See links to NCBI TaxBrowser for present taxonomy
 |