Biology:Phosphoinositide 3-kinase
| Phosphatidylinositol-4,5-bisphosphate 3-kinase | |||||||||
|---|---|---|---|---|---|---|---|---|---|
 | |||||||||
| Identifiers | |||||||||
| Symbol | PI3K | ||||||||
| Pfam | PF00454 | ||||||||
| InterPro | IPR000403 | ||||||||
| SMART | SM00146 | ||||||||
| PROSITE | PDOC00710 | ||||||||
| SCOP2 | 3gmm / SCOPe / SUPFAM | ||||||||
| OPM superfamily | 265 | ||||||||
| OPM protein | 3ml9 | ||||||||
| |||||||||
| Phosphoinositide 3-kinase | |||||||||
|---|---|---|---|---|---|---|---|---|---|
| Identifiers | |||||||||
| EC number | 2.7.1.137 | ||||||||
| CAS number | 115926-52-8 | ||||||||
| Databases | |||||||||
| IntEnz | IntEnz view | ||||||||
| BRENDA | BRENDA entry | ||||||||
| ExPASy | NiceZyme view | ||||||||
| KEGG | KEGG entry | ||||||||
| MetaCyc | metabolic pathway | ||||||||
| PRIAM | profile | ||||||||
| PDB structures | RCSB PDB PDBe PDBsum | ||||||||
| |||||||||
Phosphoinositide 3-kinases (PI3Ks), also called phosphatidylinositol 3-kinases, are a family of enzymes involved in cellular functions such as cell growth, proliferation, differentiation, motility, survival and intracellular trafficking, which in turn are involved in cancer.
PI3Ks are a family of related intracellular signal transducer enzymes capable of phosphorylating the 3 position hydroxyl group of the inositol ring of phosphatidylinositol (PtdIns).[1] The pathway, with oncogene PIK3CA and tumor suppressor gene PTEN, is implicated in the sensitivity of cancer tumors to insulin and IGF1, and in calorie restriction.[2][3]
Discovery
The discovery of PI3Ks by Lewis Cantley and colleagues began with their identification of a previously unknown phosphoinositide kinase associated with the polyoma middle T protein.[4] They observed unique substrate specificity and chromatographic properties of the products of the lipid kinase, leading to the discovery that this phosphoinositide kinase had the unprecedented ability to phosphorylate phosphoinositides on the 3' position of the inositol ring.[5] Subsequently, Cantley and colleagues demonstrated that in vivo the enzyme prefers PtdIns(4,5)P2 as a substrate, producing the novel phosphoinositide PtdIns(3,4,5)P3[6] previously identified in neutrophils.[7]
Classes
The PI3K family is divided into four different classes: Class I, Class II, Class III, and Class IV. The classifications are based on primary structure, regulation, and in vitro lipid substrate specificity.[8]
Class I
Class I PI3Ks catalyze the conversion of phosphatidylinositol (4,5)-bisphosphate (PI(4,5)P2) into phosphatidylinositol (3,4,5)-trisphosphate (PI(3,4,5)P3) in vivo. While in vitro, they have also been shown to convert phosphatidylinositol (PI) into phosphatidylinositol 3-phosphate (PI3P) and phosphatidylinositol 4-phosphate (PI4P) into phosphatidylinositol (3,4)-bisphosphate (PI(3,4)P2), these reactions are strongly disfavoured in vivo.[9][10][11][12] The PI3K is activated by G protein-coupled receptors and tyrosine kinase receptors.[8]
Class I PI3Ks are heterodimeric molecules composed of a regulatory and a catalytic subunit; they are further divided between IA and IB subsets on sequence similarity. Class IA PI3Ks are composed of a heterodimer between a p110 catalytic subunit and a shorter regulatory subunit (often p85).[13] There are five variants of the regulatory subunit: the three splice variants p85α, p55α, and p50α, p85β, and p55γ. There are also three variants of the p110 catalytic subunit designated p110α, β, or δ catalytic subunit. The first three regulatory subunits are all splice variants of the same gene (Pik3r1), the other two being expressed by other genes (Pik3r2 and Pik3r3, p85β, and p55γ, respectively). The most highly expressed regulatory subunit is p85α; all three catalytic subunits are expressed by separate genes (Pik3ca, Pik3cb, and Pik3cd for p110α, p110β, and p110δ, respectively). The first two p110 isoforms (α and β) are expressed in all cells, but p110δ is expressed primarily in leukocytes, and it has been suggested that it evolved in parallel with the adaptive immune system. The regulatory p101 and catalytic p110γ subunits comprise the class IB PI3Ks and are encoded by a single gene each (Pik3cg for p110γ and Pik3r5 for p101).
The p85 subunits contain SH2 and SH3 domains (Online Mendelian Inheritance in Man (OMIM) 171833). The SH2 domains bind preferentially to phosphorylated tyrosine residues in the amino acid sequence context Y-X-X-M.[14][15]
Classes II and III
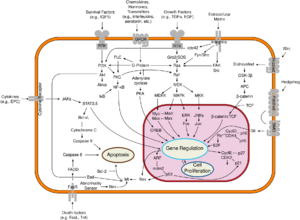
Class II and III PI3Ks are differentiated from the Class I by their structure and function. The distinct feature of Class II PI3Ks is the C-terminal C2 domain. This domain lacks critical Asp residues to coordinate binding of Ca2+, which suggests class II PI3Ks bind lipids in a Ca2+-independent manner.
Class II comprises three catalytic isoforms (C2α, C2β, and C2γ), but, unlike Classes I and III, no regulatory proteins. Class II catalyse the production of PI(3)P from PI and PI(3,4)P2 from PI(4)P; however, little is known about their role in immune cells. PI(3,4)P2 has, however, been shown to play a role in the invagination phase of clathrin-mediated endocytosis.[16] C2α and C2β are expressed through the body, but expression of C2γ is limited to hepatocytes.
Class III PI3Ks produce only PI(3)P from PI [8] but are more similar to Class I in structure, as they exist as heterodimers of a catalytic (Vps34) and a regulatory (Vps15/p150) subunits. Class III seems to be primarily involved in the trafficking of proteins and vesicles. There is, however, evidence to show that they are able to contribute to the effectiveness of several process important to immune cells, not least phagocytosis.
Class IV
A group of more distantly related enzymes is sometimes referred to as class IV PI3Ks. It is composed of ataxia telangiectasia mutated (ATM), ataxia telangiectasia and Rad3 related (ATR), DNA-dependent protein kinase (DNA-PK) and mammalian target of rapamycin (mTOR). They are protein serine/threonine kinases.
Human genes
| group | gene | protein | aliases | EC number |
|---|---|---|---|---|
| class 1 catalytic | PIK3CA | PI3K, catalytic, alpha polypeptide | p110-α | 2.7.1.153 |
| PIK3CB | PI3K, catalytic, beta polypeptide | p110-β | ||
| PIK3CG | PI3K, catalytic, gamma polypeptide | p110-γ | ||
| PIK3CD | PI3K, catalytic, delta polypeptide | p110-δ | ||
| class 1 regulatory | PIK3R1 | PI3K, regulatory subunit 1 (alpha) | p85-α, p55-α, p50-α (splice variants) | N/A |
| PIK3R2 | PI3K, regulatory subunit 2 (beta) | p85-β | ||
| PIK3R3 | PI3K, regulatory subunit 3 (gamma) | p55-γ | ||
| PIK3R4 | PI3K, regulatory subunit 4 | p150 | ||
| PIK3R5 | PI3K, regulatory subunit 5 | p101 | ||
| PIK3R6 | PI3K, regulatory subunit 6 | p87 | ||
| class 2 | PIK3C2A | PI3K, class 2, alpha polypeptide | PI3K-C2α | 2.7.1.154 |
| PIK3C2B | PI3K, class 2, beta polypeptide | PI3K-C2β | ||
| PIK3C2G | PI3K, class 2, gamma polypeptide | PI3K-C2γ | ||
| class 3 | PIK3C3 | PI3K, class 3 | Vps34 | 2.7.1.137 |
Mechanism
The various 3-phosphorylated phosphoinositides that are produced by PI3Ks (PtdIns3P, PtdIns(3,4)P2, PtdIns(3,5)P2, and PtdIns(3,4,5)P3) function in a mechanism by which an assorted group of signalling proteins, containing PX domains, pleckstrin homology domains (PH domains), FYVE domains or other phosphoinositide-binding domains, are recruited to various cellular membranes.
Function
PI3Ks have been linked to an extraordinarily diverse group of cellular functions, including cell growth, proliferation, differentiation, motility, survival and intracellular trafficking. Many of these functions relate to the ability of class I PI3Ks to activate protein kinase B (PKB, aka Akt) as in the PI3K/AKT/mTOR pathway. The p110δ and p110γ isoforms regulate different aspects of immune responses. PI3Ks are also a key component of the insulin signaling pathway. Hence there is great interest in the role of PI3K signaling in diabetes mellitus. PI3K is also involved in interleukin signalling (IL4)[citation needed]
Mechanism
The pleckstrin homology domain of AKT binds directly to PtdIns(3,4,5)P3 and PtdIns(3,4)P2, which are produced by activated PI3Ks.[17] Since PtdIns(3,4,5)P3 and PtdIns(3,4)P2 are restricted to the plasma membrane, this results in translocation of AKT to the plasma membrane. Likewise, the phosphoinositide-dependent kinase-1 (PDK1 or, rarely referred to as PDPK1) also contains a pleckstrin homology domain that binds directly to PtdIns(3,4,5)P3 and PtdIns(3,4)P2, causing it to also translocate to the plasma membrane upon PI3K activation. The interaction of activated PDK1 and AKT allows AKT to become phosphorylated by PDK1 on threonine 308, leading to partial activation of AKT. Full activation of AKT occurs upon phosphorylation of serine 473 by the TORC2 complex of the mTOR protein kinase.
The PI3K/AKT pathway has been shown to be required for an extremely diverse array of cellular activities - most notably cellular proliferation and survival. For example, it was shown to be involved in the protection of astrocytes from ceramide-induced apoptosis.[18]
Many other proteins have been identified that are regulated by PtdIns(3,4,5)P3, including Bruton's tyrosine kinase (BTK), General Receptor for Phosphoinositides-1 (GRP1), and the O-linked N-acetylglucosamine (O-GlcNAc) transferase.
PtdIns(3,4,5)P3 also activates guanine‐nucleotide exchange factors (GEFs) that activate the GTPase Rac1,[19] leading to actin polymerization and cytoskeletal rearrangement.[20]
Cancers
The class IA PI3K p110α is mutated in many cancers. Many of these mutations cause the kinase to be more active. It is the single most mutated kinase in glioblastoma, the most malignant primary brain tumor.[21] The PtdIns(3,4,5)P3 phosphatase PTEN that antagonises PI3K signaling is absent from many tumours. In addition, the epidermal growth factor receptor EGFR that functions upstream of PI3K is mutationally activated or overexpressed in cancer.[21][22] Hence, PI3K activity contributes significantly to cellular transformation and the development of cancer. It has been shown that malignant B cells maintain a "tonic" activity of PI3K/Akt axis via upregulation of an adaptor protein GAB1, and this also allows B cells to survive targeted therapy with BCR inhibitors.[citation needed]
Learning and memory
PI3Ks have also been implicated in long-term potentiation (LTP). Whether they are required for the expression or the induction of LTP is still debated. In mouse hippocampal CA1 neurons, certain PI3Ks are complexed with AMPA receptors and compartmentalized at the postsynaptic density of glutamatergic synapses.[23] PI3Ks are phosphorylated upon NMDA receptor-dependent CaMKII activity,[24] and it then facilitates the insertion of AMPA-R GluR1 subunits into the plasma membrane. This suggests that PI3Ks are required for the expression of LTP. Furthermore, PI3K inhibitors abolished the expression of LTP in rat hippocampal CA1, but do not affect its induction.[25] Notably, the dependence of late-phase LTP expression on PI3Ks seems to decrease over time.[26]
However, another study found that PI3K inhibitors suppressed the induction, but not the expression, of LTP in mouse hippocampal CA1.[27] The PI3K pathway also recruits many other proteins downstream, including mTOR,[28] GSK3β,[29] and PSD-95.[28] The PI3K-mTOR pathway leads to the phosphorylation of p70S6K, a kinase that facilitates translational activity,[30][31] further suggesting that PI3Ks are required for the protein-synthesis phase of LTP induction instead.
PI3Ks interact with the insulin receptor substrate (IRS) to regulate glucose uptake through a series of phosphorylation events.
PI 3-kinases as protein kinases
Many PI3Ks appear to have a serine/threonine kinase activity in vitro; however, it is unclear whether this has any role in vivo.[citation needed]
Inhibition
All PI3Ks are inhibited by the drugs wortmannin and LY294002, although certain members of the class II PI3K family show decreased sensitivity. Wortmannin shows better efficiency than LY294002 on the hotspot mutation positions (GLU542, GLU545, and HIS1047)[32][33]
PI3K inhibitors as therapeutics
As wortmannin and LY294002 are broad-range inhibitors of PI3Ks and a number of unrelated proteins at higher concentrations, they are too toxic to be used as therapeutics.[citation needed] A number of pharmaceutical companies have thus developed PI3K isoform-specific inhibitors. As of January 2019, three PI3K inhibitors are approved by the FDA for routine clinical use in humans: the PIK3CD inhibitor idelalisib (July 2014, NDA 206545), the dual PIK3CA and PIK3CD inhibitor copanlisib (September 2017, NDA 209936), and the dual PIK3CD and PIK3CG inhibitor duvelisib (September 2018, NDA 211155). Co-targeted inhibition of the pathway with other pathways such as MAPK or PIM has been highlighted as a promising anti-cancer therapeutic strategy, which could offer benefit over the monotherapeutic approach by circumventing compensatory signalling, slowing the development of resistance and potentially allowing reduction of dosing.[34][35][36][37][38]
See also
- PI3K/AKT/mTOR pathway
References
- ↑ "myo-inositol". http://www.chem.qmul.ac.uk/iupac/cyclitol/myo.html.
- ↑ Giese N (2009). "Cell pathway on overdrive prevents cancer response to dietary restriction". PhysOrg.com. http://www.physorg.com/news156000728.html.
- ↑ "Tumours with PI3K activation are resistant to dietary restriction". Nature 458 (7239): 725–31. April 2009. doi:10.1038/nature07782. PMID 19279572. Bibcode: 2009Natur.458..725K.
- ↑ "Association of phosphatidylinositol kinase activity with polyoma middle-T competent for transformation". Nature 315 (6016): 239–42. 1985. doi:10.1038/315239a0. PMID 2987699. Bibcode: 1985Natur.315..239W.
- ↑ "Type I phosphatidylinositol kinase makes a novel inositol phospholipid, phosphatidylinositol-3-phosphate". Nature 332 (6165): 644–6. April 1988. doi:10.1038/332644a0. PMID 2833705. Bibcode: 1988Natur.332..644W.
- ↑ "PDGF-dependent tyrosine phosphorylation stimulates production of novel polyphosphoinositides in intact cells". Cell 57 (1): 167–75. April 1989. doi:10.1016/0092-8674(89)90182-7. PMID 2467744.
- ↑ "An inositol tetrakisphosphate-containing phospholipid in activated neutrophils". Nature 334 (6180): 353–6. July 1988. doi:10.1038/334353a0. PMID 3393226. Bibcode: 1988Natur.334..353T.
- ↑ 8.0 8.1 8.2 "Signalling through phosphoinositide 3-kinases: the lipids take centre stage". Current Opinion in Cell Biology 11 (2): 219–25. April 1999. doi:10.1016/S0955-0674(99)80029-5. PMID 10209156.
- ↑ "The PI3K Pathway in Human Disease". Cell 170 (4): 605–635. August 2017. doi:10.1016/j.cell.2017.07.029. PMID 28802037.
- ↑ "Classes of phosphoinositide 3-kinases at a glance". Journal of Cell Science 127 (Pt 5): 923–8. March 2014. doi:10.1242/jcs.093773. PMID 24587488.
- ↑ "PI3K signalling: the path to discovery and understanding". Nature Reviews. Molecular Cell Biology 13 (3): 195–203. February 2012. doi:10.1038/nrm3290. PMID 22358332.
- ↑ "Signaling by the phosphoinositide 3-kinase family in immune cells". Annual Review of Immunology 31 (2): 675–704. January 2013. doi:10.1146/annurev-immunol-032712-095946. PMID 23330955.
- ↑ "Purification and characterization of phosphoinositide 3-kinase from rat liver". The Journal of Biological Chemistry 265 (32): 19704–11. November 1990. doi:10.1016/S0021-9258(17)45429-9. PMID 2174051.
- ↑ "SH2 domains recognize specific phosphopeptide sequences". Cell 72 (5): 767–78. March 1993. doi:10.1016/0092-8674(93)90404-E. PMID 7680959.
- ↑ "Genetic analysis of a phosphatidylinositol 3-kinase SH2 domain reveals determinants of specificity". Molecular and Cellular Biology 14 (9): 5929–38. September 1994. doi:10.1128/MCB.14.9.5929. PMID 8065326.
- ↑ "Phosphoinositides in endocytosis". Biochimica et Biophysica Acta (BBA) - Molecular and Cell Biology of Lipids 1851 (6): 794–804. June 2015. doi:10.1016/j.bbalip.2014.09.014. PMID 25264171.
- ↑ "Direct regulation of the Akt proto-oncogene product by phosphatidylinositol-3,4-bisphosphate". Science 275 (5300): 665–8. January 1997. doi:10.1126/science.275.5300.665. PMID 9005852.
- ↑ "Cannabinoids protect astrocytes from ceramide-induced apoptosis through the phosphatidylinositol 3-kinase/protein kinase B pathway". The Journal of Biological Chemistry 277 (39): 36527–33. September 2002. doi:10.1074/jbc.M205797200. PMID 12133838.
- ↑ "Phosphoinositide 3-kinase-dependent activation of Rac". FEBS Letters 546 (1): 93–7. July 2003. doi:10.1016/s0014-5793(03)00454-x. PMID 12829242.
- ↑ "Rho GTPases: biochemistry and biology". Annual Review of Cell and Developmental Biology 21: 247–69. 2005. doi:10.1146/annurev.cellbio.21.020604.150721. PMID 16212495.
- ↑ 21.0 21.1 "Mutational profiling of kinases in glioblastoma". BMC Cancer 14: 718. September 2014. doi:10.1186/1471-2407-14-718. PMID 25256166.
- ↑ "Recent advances in the molecular understanding of glioblastoma". Journal of Neuro-Oncology 108 (1): 11–27. May 2012. doi:10.1007/s11060-011-0793-0. PMID 22270850.
- ↑ "Activation of PI3-kinase is required for AMPA receptor insertion during LTP of mEPSCs in cultured hippocampal neurons". Neuron 38 (4): 611–24. May 2003. doi:10.1016/S0896-6273(03)00228-9. PMID 12765612.
- ↑ "Calmodulin activates phosphatidylinositol 3-kinase". The Journal of Biological Chemistry 272 (45): 28183–6. November 1997. doi:10.1074/jbc.272.45.28183. PMID 9353264.
- ↑ "Phosphatidylinositol 3-kinase is required for the expression but not for the induction or the maintenance of long-term potentiation in the hippocampal CA1 region". The Journal of Neuroscience 22 (9): 3359–65. May 2002. doi:10.1523/JNEUROSCI.22-09-03359.2002. PMID 11978812.
- ↑ "Involvement of multiple phosphatidylinositol 3-kinase-dependent pathways in the persistence of late-phase long term potentiation expression". Neuroscience 137 (3): 833–41. February 2006. doi:10.1016/j.neuroscience.2005.10.012. PMID 16326012.
- ↑ "Phosphatidylinositol 3-kinase regulates the induction of long-term potentiation through extracellular signal-related kinase-independent mechanisms". The Journal of Neuroscience 23 (9): 3679–88. May 2003. doi:10.1523/JNEUROSCI.23-09-03679.2003. PMID 12736339.
- ↑ 28.0 28.1 "Phosphatidylinositol 3-kinase activation is required for stress protocol-induced modification of hippocampal synaptic plasticity". The Journal of Biological Chemistry 283 (5): 2631–43. February 2008. doi:10.1074/jbc.M706954200. PMID 18057005.
- ↑ "LTP inhibits LTD in the hippocampus via regulation of GSK3beta". Neuron 53 (5): 703–17. March 2007. doi:10.1016/j.neuron.2007.01.029. PMID 17329210. http://www.hal.inserm.fr/inserm-00776885.
- ↑ "Signalling through the lipid products of phosphoinositide-3-OH kinase". Nature 387 (6634): 673–6. June 1997. doi:10.1038/42648. PMID 9192891. Bibcode: 1997Natur.387..673T.
- ↑ "Time-restricted role for dendritic activation of the mTOR-p70S6K pathway in the induction of late-phase long-term potentiation in the CA1". Proceedings of the National Academy of Sciences of the United States of America 100 (24): 14368–73. November 2003. doi:10.1073/pnas.2336098100. PMID 14623952. Bibcode: 2003PNAS..10014368C.
- ↑ Investigating the Inhibitory Effect of Wortmannin in the Hotspot Mutation at Codon 1047 of PIK3CA Kinase Domain: A Molecular Docking and Molecular Dynamics Approach. 102. 2016-01-01. 267–97. doi:10.1016/bs.apcsb.2015.09.008. ISBN 9780128047958.
- ↑ "Deciphering the impact of somatic mutations in exon 20 and exon 9 of PIK3CA gene in breast tumors among Indian women through molecular dynamics approach". Journal of Biomolecular Structure & Dynamics 34 (1): 29–41. 2016-01-02. doi:10.1080/07391102.2015.1007483. PMID 25679319.
- ↑ "Current perspectives on targeting PIM kinases to overcome mechanisms of drug resistance and immune evasion in cancer". Pharmacology & Therapeutics 207: 107454. March 2020. doi:10.1016/j.pharmthera.2019.107454. PMID 31836451. https://discovery.ucl.ac.uk/id/eprint/10088376/1/1-s2.0-S0163725819302062-main.pdf.
- ↑ "PIM kinase inhibition: co-targeted therapeutic approaches in prostate cancer". Signal Transduction and Targeted Therapy 5: 7. 2020. doi:10.1038/s41392-020-0109-y. PMID 32025342.
- ↑ "Development and characterisation of a panel of phosphatidylinositide 3-kinase - mammalian target of rapamycin inhibitor resistant lung cancer cell lines". Scientific Reports 8 (1): 1652. January 2018. doi:10.1038/s41598-018-19688-1. PMID 29374181. Bibcode: 2018NatSR...8.1652H.
- ↑ "Strategic targeting of the PI3K-NFκB axis in cisplatin-resistant NSCLC". Cancer Biology & Therapy 15 (10): 1367–77. October 2014. doi:10.4161/cbt.29841. PMID 25025901.
- ↑ "Strategies for co-targeting the PI3K/AKT/mTOR pathway in NSCLC". Cancer Treatment Reviews 40 (3): 445–56. April 2014. doi:10.1016/j.ctrv.2013.08.006. PMID 24055012.
Further reading
- "Synthesis and function of 3-phosphorylated inositol lipids". Annual Review of Biochemistry 70: 535–602. 2001. doi:10.1146/annurev.biochem.70.1.535. PMID 11395417. [1]
- "PI3K signaling maintains c-myc expression to regulate transcription of E2F1 in pancreatic cancer cells". Molecular Carcinogenesis 48 (12): 1149–58. December 2009. doi:10.1002/mc.20569. PMID 19603422.
- "Form and flexibility in phosphoinositide 3-kinases". Biochemical Society Transactions 37 (Pt 4): 615–26. August 2009. doi:10.1042/BST0370615. PMID 19614567.
- "Regulation of mRNA export by the PI3 kinase/AKT signal transduction pathway". Molecular Biology of the Cell 24 (8): 1208–21. April 2013. doi:10.1091/mbc.E12-06-0450. PMID 23427269.
External links
- Eukaryotic Linear Motif resource motif class MOD_PIKK_1
- Proteopedia Phosphoinositide_3-Kinases to explore the structure in interactive 3D
- PI-3+Kinase at the US National Library of Medicine Medical Subject Headings (MeSH)
- PI3K/Akt Signaling Pathway
 |

