Biology:Small nucleolar RNA
In molecular biology, small nucleolar RNAs (snoRNAs) are a class of small RNA molecules that primarily guide chemical modifications of other RNAs, mainly ribosomal RNAs, transfer RNAs and small nuclear RNAs. There are two main classes of snoRNA, the C/D box snoRNAs, which are associated with methylation, and the H/ACA box snoRNAs, which are associated with pseudouridylation. SnoRNAs are commonly referred to as guide RNAs but should not be confused with the guide RNAs that direct RNA editing in trypanosomes or the guide RNAs (gRNAs) used by Cas9 for CRISPR gene editing.
snoRNA guided modifications
After transcription, nascent rRNA molecules (termed pre-rRNA) undergo a series of processing steps to generate the mature rRNA molecule. Prior to cleavage by exo- and endonucleases, the pre-rRNA undergoes a complex pattern of nucleoside modifications. These include methylations and pseudouridylations, guided by snoRNAs.
- Methylation is the attachment or substitution of a methyl group onto various substrates. The rRNA of humans contain approximately 115 methyl group modifications. The majority of these are 2′O-ribose-methylations (where the methyl group is attached to the ribose group).[1]
- Pseudouridylation is the conversion (isomerisation) of the nucleoside uridine to a different isomeric form pseudouridine (Ψ). This modification consists of a 180º rotation of the uridine base around its glycosyl bond to the ribose of the RNA backbone. After this rotation, the nitrogenous base contributes a carbon atom to the glycosyl bond instead of the usual nitrogen atom. The beneficial aspect of this modification is the additional hydrogen-bond donor available on the base. While uridine makes two hydrogen-bonds with its Watson-Crick base pair, adenine, pseudouridine is capable of making three hydrogen bonds. When pseudouridine is base-paired with adenine, it can also make another hydrogen bond, allowing the complexity of the mature rRNA structure to take form. The free hydrogen-bond donor often forms a bond with a base that is distant from itself, creating the tertiary structure that rRNA must have to be functional. Mature human rRNAs contain approximately 95 Ψ modifications.[1]
snoRNP
Each snoRNA molecule acts as a guide for only one (or two) individual modifications in a target RNA.[2] In order to carry out modification, each snoRNA associates with at least four core proteins in an RNA/protein complex referred to as a small nucleolar ribonucleoprotein particle (snoRNP).[3] The proteins associated with each RNA depend on the type of snoRNA molecule (see snoRNA guide families below). The snoRNA molecule contains an antisense element (a stretch of 10–20 nucleotides), which are base complementary to the sequence surrounding the base (nucleotide) targeted for modification in the pre-RNA molecule. This enables the snoRNP to recognise and bind to the target RNA. Once the snoRNP has bound to the target site, the associated proteins are in the correct physical location to catalyse the chemical modification of the target base.[4]
snoRNA guide families
The two different types of rRNA modification (methylation and pseudouridylation) are directed by two different families of snoRNAs. These families of snoRNAs are referred to as antisense C/D box and H/ACA box snoRNAs based on the presence of conserved sequence motifs in the snoRNA. There are exceptions, but as a general rule C/D box members guide methylation and H/ACA members guide pseudouridylation. The members of each family may vary in biogenesis, structure, and function, but each family is classified by the following generalised characteristics. For more detail, see review.[5] SnoRNAs are classified under small nuclear RNA in MeSH. The HGNC, in collaboration with snoRNABase and experts in the field, has approved unique names for human genes that encode snoRNAs.[6]
C/D box
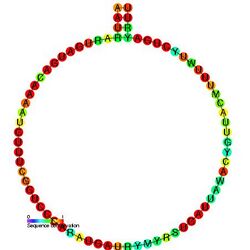
C/D box snoRNAs contain two short conserved sequence motifs, C (RUGAUGA) and D (CUGA), located near the 5′ and 3′ ends of the snoRNA, respectively. Short regions (~ 5 nucleotides) located upstream of the C box and downstream of the D box are usually base complementary and form a stem-box structure, which brings the C and D box motifs into close proximity. This stem-box structure has been shown to be essential for correct snoRNA synthesis and nucleolar localization.[7] Many C/D box snoRNA also contain an additional less-well-conserved copy of the C and D motifs (referred to as C' and D') located in the central portion of the snoRNA molecule. A conserved region of 10–21 nucleotides upstream of the D box is complementary to the methylation site of the target RNA and enables the snoRNA to form an RNA duplex with the RNA.[8] The nucleotide to be modified in the target RNA is usually located at the 5th position upstream from the D box (or D' box).[9][10] C/D box snoRNAs associate with four evolutionary conserved and essential proteins—fibrillarin (Nop1p), NOP56, NOP58, and SNU13 (15.5-kD protein in eukaryotes; its archaeal homolog is L7Ae)—which make up the core C/D box snoRNP.[5]
There exists a eukaryotic C/D box snoRNA (snoRNA U3) that has not been shown to guide 2′-O-methylation. Instead, it functions in rRNA processing by directing pre-rRNA cleavage.
H/ACA box
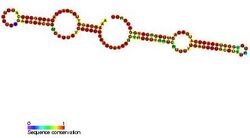
H/ACA box snoRNAs have a common secondary structure consisting of a two hairpins and two single-stranded regions termed a hairpin-hinge-hairpin-tail structure.[5] H/ACA snoRNAs also contain conserved sequence motifs known as H box (consensus ANANNA) and the ACA box (ACA). Both motifs are usually located in the single-stranded regions of the secondary structure. The H motif is located in the hinge and the ACA motif is located in the tail region; 3 nucleotides from the 3′ end of the sequence.[11] The hairpin regions contain internal bulges known as recognition loops in which the antisense guide sequences (bases complementary to the target sequence) are located. These guide sequences essentially mark the location of the uridine on the target rRNA that is going to be modified. This recognition sequence is bipartite (constructed from the two different arms of the loop region) and forms complex pseudo-knots with the target RNA. H/ACA box snoRNAs associate with four evolutionary conserved and essential proteins—dyskerin (Cbf5p), GAR1, NHP2, and NOP10—which make up the core of the H/ACA box snoRNP.[5] Dyskerin is likely the catalytic component of the ribonucleoprotein (RNP) complex because it possesses several conserved pseudouridine synthase sequences, and is closely related to the pseudouridine synthase that modifies uridine in tRNA. In lower eukaryotic cells such as trypanosomes, similar RNAs exist in the form of single hairpin structure and an AGA box instead of ACA box at the 3′ end of the RNA.[12] Like Trypanosomes, Entamoeba histolytica has mix population of single hairpin as well as double hairpin H/ACA box snoRNAs. It was reported that there occurred processing of the double hairpin H/ACA box snoRNA to the single hairpin snoRNAs however, unlike trypanosomes, it has a regular ACA motif at 3′ tail.[19]
The RNA component of human telomerase (hTERC) contains an H/ACA domain for pre-RNP formation and nucleolar localization of the telomerase RNP itself.[13] The H/ACA snoRNP has been implicated in the rare genetic disease dyskeratosis congenita (DKC) due to its affiliation with human telomerase. Mutations in the protein component of the H/ACA snoRNP result in a reduction in physiological TERC levels. This has been strongly correlated with the pathology behind DKC, which seems to be primarily a disease of poor telomere maintenance.
Composite H/ACA and C/D box
An unusual guide snoRNA U85 that functions in both 2′-O-ribose methylation and pseudouridylation of small nuclear RNA (snRNA) U5 has been identified.[14] This composite snoRNA contains both C/D and H/ACA box domains and associates with the proteins specific to each class of snoRNA (fibrillarin and Gar1p, respectively). More composite snoRNAs have now been characterised.[15]
These composite snoRNAs have been found to accumulate in a subnuclear organelle called the Cajal body and are referred to as small Cajal body-specific RNAs (scaRNAs). This is in contrast to the majority of C/D box or H/ACA box snoRNAs, which localise to the nucleolus. These Cajal body specific RNAs are proposed to be involved in the modification of RNA polymerase II transcribed spliceosomal RNAs U1, U2, U4, U5 and U12.[15] Not all snoRNAs that have been localised to Cajal bodies are composite C/D and H/ACA box snoRNAs.
Orphan snoRNAs
The targets for newly identified snoRNAs are predicted on the basis of sequence complementarity between putative target RNAs and the antisense elements or recognition loops in the snoRNA sequence. However, there are increasing numbers of 'orphan' guides without any known RNA targets, which suggests that there might be more proteins or transcripts involved in rRNA than previously and/or that some snoRNAs have different functions not concerning rRNA.[16][17] There is evidence that some of these orphan snoRNAs regulate alternatively spliced transcripts.[18] For example, it appears that the C/D box snoRNA SNORD115 regulates the alternative splicing of the serotonin 2C receptor mRNA via a conserved region of complementarity.[19][20] Another C/D box snoRNA, SNORD116, that resides in the same cluster as SNORD115 has been predicted to have 23 possible targets within protein coding genes using a bioinformatic approach. Of these, a large fraction were found to be alternatively spliced, suggesting a role of SNORD116 in the regulation of alternative splicing.[21]
More recently, SNORD90 has been suggested to be able to guide N6-methyladenosine (m6A) modifications onto target RNA transcripts.[22] More specifically, Lin et al. demonstrated that SNORD90 can reduce the expression of neuregulin 3 (NRG3).[22]
Target modifications
The precise effect of the methylation and pseudouridylation modifications on the function of the mature RNAs is not yet known. The modifications do not appear to be essential but are known to subtly enhance the RNA folding and interaction with ribosomal proteins. In support of their importance, target site modifications are exclusively located within conserved and functionally important domains of the mature RNA and are commonly conserved among distant eukaryotes.[5] A novel method, Nm-REP-seq, was developed for enriching 2'-O-Methylations guided by C/D snoRNAs by using RNA exoribonuclease (Mycoplasma genitalium RNase R, MgR) and periodate oxidation reactivity to eliminate 2'-hydroxylated (2'-OH) nucleosides.[23]
- 2′-O-methylated ribose causes an increase in the 3′-endo conformation
- Pseudouridine (psi/Ψ) adds another option for H-bonding.
- Heavily methylated RNA is protected from hydrolysis. rRNA acts as a ribozyme by catalyzing its own hydrolysis and splicing.
Genomic organisation
SnoRNAs are located diversely in the genome. The majority of vertebrate snoRNA genes are encoded in the introns of genes encoding proteins involved in ribosome synthesis or translation, and are synthesized by RNA polymerase II. SnoRNAs are also shown to be located in intergenic regions, ORFs of protein coding genes, and UTRs.[24] SnoRNAs can also be transcribed from their own promoters by RNA polymerase II or III.
Imprinted loci
In the human genome, there are at least two examples where C/D box snoRNAs are found in tandem repeats within imprinted loci. These two loci (14q32 on chromosome 14 and 15q11q13 on chromosome 15) have been extensively characterised, and in both regions multiple snoRNAs have been found located within introns in clusters of closely related copies.
In 15q11q13, five different snoRNAs have been identified (SNORD64, SNORD107, SNORD108, SNORD109 (two copies), SNORD116 (29 copies) and SNORD115 (48 copies). Loss of the 29 copies of SNORD116 (HBII-85) from this region has been identified as a cause of Prader-Willi syndrome[25][26][27][28] whereas gain of additional copies of SNORD115 has been linked to autism.[29][30][31]
Region 14q32 contains repeats of two snoRNAs SNORD113 (9 copies) and SNORD114 (31 copies) within the introns of a tissue-specific ncRNA transcript (MEG8). The 14q32 domain has been shown to share common genomic features with the imprinted 15q11-q13 loci and a possible role for tandem repeats of C/D box snoRNAs in the evolution or mechanism of imprinted loci has been suggested.[32][33]
Other functions
snoRNAs can function as miRNAs. It has been shown that human ACA45 is a bona fide snoRNA that can be processed into a 21-nucleotides-long mature miRNA by the RNAse III family endoribonuclease dicer.[34] This snoRNA product has previously been identified as mmu-miR-1839 and was shown to be processed independently from the other miRNA-generating endoribonuclease drosha.[35] Bioinformatical analyses have revealed that putatively snoRNA-derived, miRNA-like fragments occur in different organisms.[36]
Recently, it has been found that snoRNAs can have functions not related to rRNA. One such function is the regulation of alternative splicing of the trans gene transcript, which is done by the snoRNA HBII-52, which is also known as SNORD115.[19]
In November 2012, Schubert et al. revealed that specific RNAs control chromatin compaction and accessibility in Drosophila cells.[37]
In July 2023, Lin et al. showed that snoRNAs have the potential to guide other RNA modifications, specifically N6-methyladenosine, however this is subject to further investigation.[22]
References
- ↑ 1.0 1.1 "Eukaryotic ribosomal RNA: the recent excitement in the nucleotide modification problem". Chromosoma 105 (7–8): 391–400. June 1997. doi:10.1007/BF02510475. PMID 9211966.
- ↑ "Eurkaroytic Cellular DNA". RNA Purification and Analysis: Sample Preparation, Extraction, Chromatography. Weinheim: Wiley-VCH. 2009. pp. 25–26. ISBN 978-3-527-62720-2. https://books.google.com/books?id=qoshFRNUJS8C&pg=PA25. Retrieved 28 September 2020.
- ↑ "The snoRNPs and Related Machines: Ancient Devices That Mediate Maturation of rRNA and Other RNAs" (in en). Madame Curie Bioscience Database. Austin, Texas: Landes Bioscience. 2013. https://www.ncbi.nlm.nih.gov/books/NBK6107/. Retrieved 28 September 2020.
- ↑ "Box C/D snoRNP Autoregulation by a cis-Acting snoRNA in the NOP56 Pre-mRNA". Molecular Cell 72 (1): 99–111.e5. October 2018. doi:10.1016/j.molcel.2018.08.017. PMID 30220559.
- ↑ 5.0 5.1 5.2 5.3 5.4 "The expanding snoRNA world". Biochimie 84 (8): 775–790. August 2002. doi:10.1016/S0300-9084(02)01402-5. PMID 12457565.
- ↑ "Naming 'junk': human non-protein coding RNA (ncRNA) gene nomenclature". Human Genomics 5 (2): 90–98. January 2011. doi:10.1186/1479-7364-5-2-90. PMID 21296742.
- ↑ "The snoRNA box C/D motif directs nucleolar targeting and also couples snoRNA synthesis and localization". The EMBO Journal 17 (13): 3747–3757. July 1998. doi:10.1093/emboj/17.13.3747. PMID 9649444.
- ↑ "Sequence and structural elements of methylation guide snoRNAs essential for site-specific ribose methylation of pre-rRNA". The EMBO Journal 17 (3): 797–807. February 1998. doi:10.1093/emboj/17.3.797. PMID 9451004.
- ↑ "Targeted ribose methylation of RNA in vivo directed by tailored antisense RNA guides". Nature 383 (6602): 732–735. October 1996. doi:10.1038/383732a0. PMID 8878486. Bibcode: 1996Natur.383..732C.
- ↑ "Site-specific ribose methylation of preribosomal RNA: a novel function for small nucleolar RNAs". Cell 85 (7): 1077–1088. June 1996. doi:10.1016/S0092-8674(00)81308-2. PMID 8674114.
- ↑ "The family of box ACA small nucleolar RNAs is defined by an evolutionarily conserved secondary structure and ubiquitous sequence elements essential for RNA accumulation". Genes & Development 11 (7): 941–956. April 1997. doi:10.1101/gad.11.7.941. PMID 9106664.
- ↑ "Identification of the first trypanosome H/ACA RNA that guides pseudouridine formation on rRNA". The Journal of Biological Chemistry 276 (43): 40313–40318. October 2001. doi:10.1074/jbc.M104488200. PMID 11483606.
- ↑ "Dyskeratosis congenita mutations in the H/ACA domain of human telomerase RNA affect its assembly into a pre-RNP". RNA 15 (2): 235–243. February 2009. doi:10.1261/rna.1354009. PMID 19095616.
- ↑ "A small nucleolar guide RNA functions both in 2′-O-ribose methylation and pseudouridylation of the U5 spliceosomal RNA". The EMBO Journal 20 (3): 541–551. February 2001. doi:10.1093/emboj/20.3.541. PMID 11157760.
- ↑ 15.0 15.1 "Cajal body-specific small nuclear RNAs: a novel class of 2′-O-methylation and pseudouridylation guide RNAs". The EMBO Journal 21 (11): 2746–2756. June 2002. doi:10.1093/emboj/21.11.2746. PMID 12032087.
- ↑ "Characterisation of the U83 and U84 small nucleolar RNAs: two novel 2′-O-ribose methylation guide RNAs that lack complementarities to ribosomal RNAs" (Free full text). Nucleic Acids Research 28 (6): 1348–1354. March 2000. doi:10.1093/nar/28.6.1348. PMID 10684929.
- ↑ "Identification and functional analysis of 20 Box H/ACA small nucleolar RNAs (snoRNAs) from Schizosaccharomyces pombe". The Journal of Biological Chemistry 280 (16): 16446–16455. April 2005. doi:10.1074/jbc.M500326200. PMID 15716270.
- ↑ "Regulation of alternative splicing by snoRNAs". Cold Spring Harbor Symposia on Quantitative Biology 71: 329–334. 2006. doi:10.1101/sqb.2006.71.024. PMID 17381313.
- ↑ 19.0 19.1 "The snoRNA HBII-52 regulates alternative splicing of the serotonin receptor 2C". Science 311 (5758): 230–232. January 2006. doi:10.1126/science.1118265. PMID 16357227. Bibcode: 2006Sci...311..230K.
- ↑ "Loss of the imprinted snoRNA mbii-52 leads to increased 5htr2c pre-RNA editing and altered 5HT2CR-mediated behaviour". Human Molecular Genetics 18 (12): 2140–2148. June 2009. doi:10.1093/hmg/ddp137. PMID 19304781.
- ↑ "snoTARGET shows that human orphan snoRNA targets locate close to alternative splice junctions". Gene 408 (1–2): 172–179. January 2008. doi:10.1016/j.gene.2007.10.037. PMID 18160232.
- ↑ 22.0 22.1 22.2 "SNORD90 induces glutamatergic signaling following treatment with monoaminergic antidepressants". eLife 12: e85316. July 2023. doi:10.7554/eLife.85316. PMID 37432876.
- ↑ "Single-base resolution mapping of 2'-O-methylation sites by an exoribonuclease-enriched chemical method". Science China Life Sciences 66 (4): 800–818. April 2023. doi:10.1007/s11427-022-2210-0. PMID 36323972.
- ↑ "Computational prediction and validation of C/D, H/ACA and Eh_U3 snoRNAs of Entamoeba histolytica". BMC Genomics 13: 390. 14 August 2012. doi:10.1186/1471-2164-13-390. PMID 22892049.
- ↑ "Deletion of the MBII-85 snoRNA gene cluster in mice results in postnatal growth retardation". PLOS Genetics 3 (12): e235. December 2007. doi:10.1371/journal.pgen.0030235. PMID 18166085.
- ↑ "Prader-Willi phenotype caused by paternal deficiency for the HBII-85 C/D box small nucleolar RNA cluster". Nature Genetics 40 (6): 719–721. June 2008. doi:10.1038/ng.158. PMID 18500341.
- ↑ "SnoRNA Snord116 (Pwcr1/MBII-85) deletion causes growth deficiency and hyperphagia in mice". PLOS ONE 3 (3): e1709. March 2008. doi:10.1371/journal.pone.0001709. PMID 18320030. Bibcode: 2008PLoSO...3.1709D.
- ↑ "Lack of Pwcr1/MBII-85 snoRNA is critical for neonatal lethality in Prader-Willi syndrome mouse models". Mammalian Genome 16 (6): 424–431. June 2005. doi:10.1007/s00335-005-2460-2. PMID 16075369.
- ↑ "Abnormal behavior in a chromosome-engineered mouse model for human 15q11–13 duplication seen in autism". Cell 137 (7): 1235–1246. June 2009. doi:10.1016/j.cell.2009.04.024. PMID 19563756.
- ↑ "Chromosome 15q11–13 abnormalities and other medical conditions in individuals with autism spectrum disorders". Psychiatric Genetics 14 (3): 131–137. September 2004. doi:10.1097/00041444-200409000-00002. PMID 15318025.
- ↑ "Copy-number variations associated with neuropsychiatric conditions". Nature 455 (7215): 919–923. October 2008. doi:10.1038/nature07458. PMID 18923514. Bibcode: 2008Natur.455..919C.
- ↑ "Identification of tandemly-repeated C/D snoRNA genes at the imprinted human 14q32 domain reminiscent of those at the Prader-Willi/Angelman syndrome region". Human Molecular Genetics 11 (13): 1527–1538. June 2002. doi:10.1093/hmg/11.13.1527. PMID 12045206.
- ↑ "Do repeated arrays of regulatory small-RNA genes elicit genomic imprinting?: Concurrent emergence of large clusters of small non-coding RNAs and genomic imprinting at four evolutionarily distinct eutherian chromosomal loci". BioEssays 33 (8): 565–573. August 2011. doi:10.1002/bies.201100032. PMID 21618561.
- ↑ "A human snoRNA with microRNA-like functions". Molecular Cell 32 (4): 519–528. November 2008. doi:10.1016/j.molcel.2008.10.017. PMID 19026782.
- ↑ "Mouse ES cells express endogenous shRNAs, siRNAs, and other Microprocessor-independent, Dicer-dependent small RNAs". Genes & Development 22 (20): 2773–2785. October 2008. doi:10.1101/gad.1705308. PMID 18923076.
- ↑ "Small RNAs derived from snoRNAs". RNA 15 (7): 1233–1240. July 2009. doi:10.1261/rna.1528909. PMID 19474147.
- ↑ "Df31 protein and snoRNAs maintain accessible higher-order structures of chromatin". Molecular Cell 48 (3): 434–444. November 2012. doi:10.1016/j.molcel.2012.08.021. PMID 23022379.
External links
- human snoRNA atlas from small RNA sequencing data
- plant snoRNA database
- snoRNAbase: human H/ACA and C/D box snoRNA database
- snoRNP Database
- The yeast snoRNA database
- human snoRNA expression pattern
- Rfam page for C/D box snoRNAs
- Rfam page for H/ACA box snoRNAs
- Rfam page for scaRNA snoRNAs
 |
