Biology:Subtilase
| Subtilase3 family | |||||||||
|---|---|---|---|---|---|---|---|---|---|
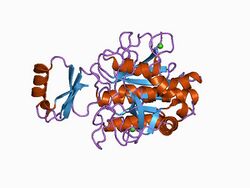 Structure of the complex formed between subtilisin Carlsberg and eglin c, an elastase inhibitor from the leech Hirudo medicinalis.[1] | |||||||||
| Identifiers | |||||||||
| Symbol | Peptidase_S8 | ||||||||
| Pfam | PF00082 | ||||||||
| InterPro | IPR000209 | ||||||||
| PROSITE | PDOC00125 | ||||||||
| MEROPS | S8 | ||||||||
| SCOP2 | 1cse / SCOPe / SUPFAM | ||||||||
| CDD | cd00306 | ||||||||
| Membranome | 546 | ||||||||
| |||||||||
Subtilases are a family of subtilisin-like serine proteases. They appear to have independently and convergently evolved an Asp/Ser/His catalytic triad, like in the trypsin serine proteases. The structure of proteins in this family shows that they have an alpha/beta fold containing a 7-stranded parallel beta sheet.
The subtilisin family is the second largest serine protease family characterised to date. Over 200 subtilases are presently known, more than 170 of which with their complete amino acid sequence.[2] Subtilase is widespread, being found in eubacteria, archaebacteria, eukaryotes and viruses.[3] The vast majority of the family are endopeptidases, although there is an exopeptidase, tripeptidyl peptidase.[3][4] Structures have been determined for several members of the subtilisin family showing that subtilisins exploit the same catalytic triad as the chymotrypsins although the residues occur in a different order (His/Asp/Ser in chymotrypsin and Asp/His/Ser in subtilisin); otherwise the structures show similarity to no other proteins.[3][4] Some subtilisins are mosaic proteins, whereas others contain N- and C-terminal extensions that show no sequence similarity to any other known protein.[3] Based on sequence homology, a subdivision into six families has been proposed.[2]
The proprotein-processing endopeptidases kexin, furin and related enzymes form a distinct subfamily known as the kexin subfamily (S8B). These preferentially cleave C-terminally to paired basic amino acids. Members of this subfamily can be identified by subtly different motifs around the active site.[3][4] Members of the kexin family, along with endopeptidases R, T and K from the yeast Tritirachium and cuticle-degrading peptidase from Metarhizium, require thiol activation. This can be attributed to the presence of Cys-173 near to the active histidine.[4] Only 1 viral member of the subtilisin family is known, a 56-kDa protease from herpes virus 1, which infects the channel catfish.[3]
Sedolisins (serine-carboxyl peptidases) are proteolytic enzymes whose fold resembles that of subtilisin; however, they are considerably larger, with the mature catalytic domains containing approximately 375 amino acids. The defining features of these enzymes are a unique catalytic triad, Ser/Glu/Asp, as well as the presence of an aspartic acid residue in the oxyanion hole. High-resolution crystal structures have now been solved for sedolisin from Pseudomonas sp. 101, as well as for kumamolisin from a thermophilic bacterium, Bacillus novo sp. MN-32. Mutations in the human gene leads to a fatal neurodegenerative disease.[5]
Human proteins containing this domain
FURIN; MBTPS1; PCSK1; PCSK2; PCSK4; PCSK5; PCSK6; PCSK7; PCSK9; TPP2;
References
- ↑ "The high-resolution X-ray crystal structure of the complex formed between subtilisin Carlsberg and eglin c, an elastase inhibitor from the leech Hirudo medicinalis. Structural analysis, subtilisin structure and interface geometry". Eur. J. Biochem. 166 (3): 673–92. August 1987. doi:10.1111/j.1432-1033.1987.tb13566.x. PMID 3301348.
- ↑ 2.0 2.1 "Subtilases: the superfamily of subtilisin-like serine proteases". Protein Sci. 6 (3): 501–523. 1997. doi:10.1002/pro.5560060301. PMID 9070434.
- ↑ 3.0 3.1 3.2 3.3 3.4 3.5 "Families of serine peptidases". Meth. Enzymol.. Methods in Enzymology 244: 19–61. 1994. doi:10.1016/0076-6879(94)44004-2. ISBN 9780121821456. PMID 7845208.
- ↑ 4.0 4.1 4.2 4.3 "Evolutionary families of peptidases". Biochem. J. 290: 205–218. 1993. doi:10.1042/bj2900205. PMID 8439290.
- ↑ "Structural and enzymatic properties of the sedolisin family of serine-carboxyl peptidases". Acta Biochim. Pol. 50 (1): 81–102. 2003. doi:10.18388/abp.2003_3716. PMID 12673349.
External links
- Eukaryotic Linear Motif resource motif class CLV_PCSK_FUR_1
- Eukaryotic Linear Motif resource motif class CLV_PCSK_PC1ET2_1
- Eukaryotic Linear Motif resource motif class CLV_PCSK_PC7_1
- Eukaryotic Linear Motif resource motif class CLV_PCSK_SKI1_1
 |
