Biology:Glycine cleavage system
| Glycine cleavage H-protein | |||||||||
|---|---|---|---|---|---|---|---|---|---|
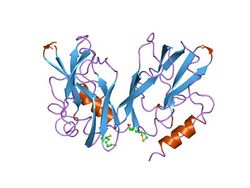 refined structures at 2 angstroms and 2.2 angstroms of the two forms of the h-protein, a lipoamide-containing protein of the glycine decarboxylase | |||||||||
| Identifiers | |||||||||
| Symbol | GCV_H | ||||||||
| Pfam | PF01597 | ||||||||
| Pfam clan | CL0105 | ||||||||
| InterPro | IPR002930 | ||||||||
| SCOP2 | 1htp / SCOPe / SUPFAM | ||||||||
| |||||||||
| Glycine cleavage T-protein, Aminomethyltransferase folate-binding domain | |||||||||
|---|---|---|---|---|---|---|---|---|---|
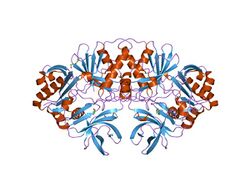 crystal structure of a component of glycine cleavage system: t-protein from pyrococcus horikoshii ot3 at 1.5 a resolution | |||||||||
| Identifiers | |||||||||
| Symbol | GCV_T | ||||||||
| Pfam | PF01571 | ||||||||
| Pfam clan | CL0289 | ||||||||
| InterPro | IPR006222 | ||||||||
| SCOP2 | 1pj5 / SCOPe / SUPFAM | ||||||||
| |||||||||
| Glycine cleavage T-protein C-terminal barrel domain | |||||||||
|---|---|---|---|---|---|---|---|---|---|
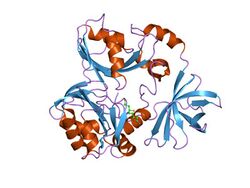 crystal structure of t-protein of the glycine cleavage system | |||||||||
| Identifiers | |||||||||
| Symbol | GCV_T_C | ||||||||
| Pfam | PF08669 | ||||||||
| InterPro | IPR013977 | ||||||||
| SCOP2 | 1pj5 / SCOPe / SUPFAM | ||||||||
| |||||||||
The glycine cleavage system (GCS) is also known as the glycine decarboxylase complex or GDC. The system is a series of enzymes that are triggered in response to high concentrations of the amino acid glycine.[1] The same set of enzymes is sometimes referred to as glycine synthase when it runs in the reverse direction to form glycine.[2] The glycine cleavage system is composed of four proteins: the T-protein, P-protein, L-protein, and H-protein. They do not form a stable complex,[3] so it is more appropriate to call it a "system" instead of a "complex". The H-protein is responsible for interacting with the three other proteins and acts as a shuttle for some of the intermediate products in glycine decarboxylation.[2] In both animals and plants, the glycine cleavage system is loosely attached to the inner membrane of the mitochondria. Mutations in this enzymatic system are linked with glycine encephalopathy.[2]
Components
| Name | EC number | Function |
|---|---|---|
| P-protein (GLDC) | EC 1.4.4.2 | glycine dehydrogenase (decarboxylating) or just glycine dehydrogenase (pyridoxal phosphate) |
| T-protein (GCST or AMT) | EC 2.1.2.10 | aminomethyltransferase |
| H-protein (GCSH) | is modified with lipoic acid and interacts with all other components in a cycle of reductive methylamination (catalysed by the P-protein), methylamine transfer (catalysed by the T-protein) and electron transfer (catalysed by the L-protein).[3] | |
| L-protein (GCSL or DLD) | EC 1.8.1.4 | known by many names, but most commonly dihydrolipoyl dehydrogenase |
Function

In plants, animals and bacteria the glycine cleavage system catalyzes the following reversible reaction:
- Glycine + H4folate + NAD+ ↔ 5,10-methylene-H4folate + CO2 + NH3 + NADH + H+
In the enzymatic reaction, H-protein activates the P-protein, which catalyzes the decarboxylation of glycine and attaches the intermediate molecule to the H-protein to be shuttled to the T-protein.[4][5] The H-protein forms a complex with the T-protein that uses tetrahydrofolate and yields ammonia and 5,10-methylenetetrahydrofolate. After interaction with the T-protein, the H-protein is left with two fully reduced thiol groups in the lipoate group.[6] The glycine protein system is regenerated when the H-protein is oxidized to regenerate the disulfide bond in the active site by interaction with the L-protein, which reduces NAD+ to NADH and H+.
When coupled to serine hydroxymethyltransferase, the glycine cleavage system overall reaction becomes:
- 2 glycine + NAD+ + H2O → serine + CO2 + NH3 + NADH + H+
In humans and most vertebrates, the glycine cleavage system is part of the most prominent glycine and serine catabolism pathway. This is due in large part to the formation 5,10-methylenetetrahydrofolate, which is one of the few C1 donors in biosynthesis.[2] In this case the methyl group derived from the catabolism of glycine can be transferred to other key molecules such as purines and methionine.

This reaction, and by extension the glycine cleavage system, is required for photorespiration in C3 plants. The glycine cleavage system takes glycine, which is created from an unwanted byproduct of the Calvin cycle, and converts it to serine which can reenter the cycle. The ammonia generated by the glycine cleavage system, is assimilated by the Glutamine synthetase-Glutamine oxoglutarate aminotransferase cycle but costs the cell one ATP and one NADPH. The upside is that one CO2 is produced for every two O2 that are mistakenly taken up by the cell, generating some value in an otherwise energy depleting cycle. Together the proteins involved in these reactions comprise about half the proteins in mitochondria from spinach and pea leaves.[3] The glycine cleavage system is constantly present in the leaves of plants, but in small amounts until they are exposed to light. During peak photosynthesis, the concentration of the glycine cleavage system increases ten-fold.[7]
In the anaerobic bacteria, Clostridium acidiurici, the glycine cleavage system runs mostly in the direction of glycine synthesis. While glycine synthesis through the cleavage system is possible due to the reversibility of the overall reaction, it is not readily seen in animals.[8][9]
Clinical significance
Glycine encephalopathy, also known as non-ketotic hyperglycinemia (NKH), is a primary disorder of the glycine cleavage system, resulting from lowered function of the glycine cleavage system causing increased levels of glycine in body fluids. The disease was first clinically linked to the glycine cleavage system in 1969.[10] Early studies showed high levels of glycine in blood, urine and cerebrospinal fluid. Initial research using carbon labeling showed decreased levels of CO2 and serine production in the liver, pointing directly to deficiencies glycine cleavage reaction.[11] Further research has shown that deletions and mutations in the 5' region of the P-protein are the major genetic causes of nonketotic hyperglycinemia. .[12] In more rare cases, a missense mutation in the genetic code of the T-protein, causing the histidine in position 42 to be mutated to arginine, was also found to result in nonketotic hypergycinemia. This specific mutation directly affected the active site of the T-protein, causing lowered efficiency of the glycine cleavage system.[13]
See also
References
- ↑ Kikuchi G (June 1973). "The glycine cleavage system: composition, reaction mechanism, and physiological significance". Mol. Cell. Biochem. 1 (2): 169–87. doi:10.1007/BF01659328. PMID 4585091.
- ↑ 2.0 2.1 2.2 2.3 Kikuchi G (2008). "The glycine cleavage system: reaction mechanism, physiological significance, and hyperglycinemia". Proc. Jpn. Acad. Ser. B Phys. Biol. Sci. 84 (7): 246–63. doi:10.2183/pjab.84.246. PMID 18941301. Bibcode: 2008PJAB...84..246K.
- ↑ 3.0 3.1 3.2 "The glycine decarboxylase system: a fascinating complex". Trends Plant Sci. 6 (4): 167–76. April 2001. doi:10.1016/S1360-1385(01)01892-1. PMID 11286922.
- ↑ "Hydrogen carrier protein from chicken liver. Purification, characterization, and role of its prosthetic group, lipoic acid, in the glycine cleavage reaction". Arch. Biochem. Biophys. 197 (2): 454–462. Oct 1979. doi:10.1016/0003-9861(79)90267-4. PMID 389161.
- ↑ "X-ray structure determination at 2.6A˚ resolution of a lipoate-containing protein. The H-protein of the glycine decraboxylase complex from pea leaves". Proc. Natl. Acad. Sci. U.S.A. 91 (11): 4850–3. May 1994. doi:10.1073/pnas.91.11.4850. PMID 8197146.
- ↑ "Mechanism of the glycine cleavage reaction. Further characterization of the intermediate attached to H-protein and of the reaction catalyzed by T-protein". J. Biol. Chem. 259 (17): 10664–8. Sep 1984. doi:10.1016/S0021-9258(18)90562-4. PMID 6469978.
- ↑ "Interaction between the component enzymes of the glycine decarboxylase mutienzyme complex". Plant Physiology 94 (4): 833–839. Oct 1990. doi:10.1104/pp.94.2.833. PMID 16667785.
- ↑ "Glycine synthase of the purinolytic bacterium Clostridium acidiurici. Purification of the glycine-CO2 exchange system". J. Biol. Chem. 259 (10): 6085–6089. May 1984. doi:10.1016/S0021-9258(20)82108-5. PMID 6427207.
- ↑ "The mitochondrial glycine cleavage system. Unique features of the glycine decarboxylation". Mol. Cell. Biochem. 45 (3): 137–49. June 1982. doi:10.1007/bf00230082. PMID 6750353.
- ↑ "Physiological significance of glycine cleavage system in human liver as revealed by the study of hyperglycinemia.". Biochem. Biophys. Res. Commun. 35 (4): 577–83. May 1969. doi:10.1016/0006-291x(69)90387-8. PMID 5788511.
- ↑ "Nonketotic hyperglycinemia: analyses of glycine cleavage system in typical and atypical cases.". J. Pediatr. 110 (6): 873–7. June 1987. doi:10.1016/S0022-3476(87)80399-2. PMID 3585602.
- ↑ "Genomic deletion within GLDC is a major cause of non-ketotic hyperglycinaemia". Journal of Medical Genetics 44 (3): e69. Mar 2007. doi:10.1136/jmg.2006.043448. PMID 17361008.
- ↑ "A missense mutation (His42Arg) in the T-protein gene from a large Israeli-Arab kindred with nonketotic hyperglycinemia". Hum. Genet. 102 (4): 430–4. April 1998. doi:10.1007/s004390050716. PMID 9600239.
it:Glicina deidrogenasi
 |
